1. Introduction
Brain health is a comprehensive concept that encompasses the multifaceted interaction of cognitive ability, mental well-being, and total neurological functioning. Unlike mental health, which focuses primarily on emotional, psychological, and social wellness, brain health offers a more holistic understanding of the multidimensional nature of the mind, brain, and interdependencies. The current healthcare systems exhibit a notable constraint in their conceptualization of brain health, predominantly emphasizing medical illnesses and disorders over the promotion of optimal cognitive functioning and well-being. This limited perspective is exemplified by the National Institutes of Health (NIH),1 which relies upon the definition of brain health as provided by the National Institute on Aging, rather than developing its comprehensive framework. Furthermore, the National Institute of Aging (NIA)'s definition of brain health, which primarily focuses on the well-being of older adults, has its limitations. It only includes four elements: cognitive health, motor function, emotional function, and tactile function. However, this narrow definition fails to capture the full breadth and significance of the brain’s capabilities and responsibilities. Specifically, it overlooks critical aspects such as social interaction,2 daily life factors such as physical activity and sleep, and overall well-being.
Measuring brain health is further complicated by variations in perception based on age, culture, ethnicity, and geography. Patient-centered assessments, which consider self-perception of cognitive function and quality of life, are essential when evaluating brain health.3 Therefore, there is a need for universally acceptable, age-appropriate, and multidimensional metrics to comprehensively measure brain health. The well-being of the brain is shaped by a blend of cognitive elements, emotional state, choices in lifestyle, and interactions within society. For instance, emotions such as anxiety and depression have a significant impact on cognitive function.4,5 Understanding and addressing these cognitive factors are vital for promoting brain health throughout life. Engaging in activities that challenge the mind, adopting a healthy lifestyle, and staying socially and intellectually active can contribute to maintaining optimal fluid intelligence and memory function. Additionally, social isolation adversely affects emotional well-being and cognitive function.6 On the other hand, lifestyle factors such as sleep, diet, and physical activity both interact with and influence these emotional, cognitive, and social functions.7 Recent advances in understanding the underlying mechanisms of sleep emphasize that it impacts a wide range of brain functions and that the consequences of sleep deprivation can be detrimental, leading to impaired memory, attention, and even neurological dysfunction.8 Meanwhile, diet and brain health have a bidirectional relationship. The changes in diet may influence psychiatric disorders through direct effects on mood, while the development of psychiatric disorders can lead to changes in eating habits.9 Physical activity on the other hand has been shown to have significant effects on human brain health regardless of age.10 It promotes improvements in brain health, including cognitive enhancement, mood regulation, pain relief, and protection against neurodegenerative diseases, primarily through the release of neurotransmitters and neurotrophins, as well as gene expression modifications. These effects have been recognized and incorporated into the Physical Activity Guidelines for Americans, which were issued by the U.S. Department of Health and Human Services (HHS).11,12 Emerging evidence also indicates that physical activity can enhance brain functions, such as memory and attention, in both children and adults.13,14 Furthermore, maintaining social connections and engaging in meaningful relationships can also have a positive impact on brain health since social interactions help prevent feelings of isolation and depression.
The diverse array of methods employed in studying brain health poses challenges for comparing studies and making recommendations for potential interventions to enhance brain health. A recent investigation15 examined over 400 distinct methods of measuring brain health, revealing that 56.1% were utilized only once. The remaining methods were classified as imaging, biological, clinical, mental health, and cognitive tests. Among these categories, only a single study incorporated outcome measures from all four distinct categories whereas approximately 32.0% of the studies encompassed measures from two categories, with the combination of imaging and cognitive measures being the most prevalent. Conversely, 63.3% of the studies solely incorporated measures from a single category, with imaging emerging as the most utilized category. The most frequently utilized imaging methods predominantly involved estimating the volume of grey16,17 and white matter in specific brain regions, notably the hippocampus and the entire brain. Additionally, methods included assessing the presence of white matter hyperintensities18 and measuring fractional anisotropy. The Trail Making Test19 and the Mini-Mental Status Examination (MMSE)20 were also among the most employed cognitive testing methods. Using cognitive testing as the sole method for evaluating brain health presents several limitations such as cost, limited sensitivity, and potential biases due to repeated use. Meanwhile, the limitations of imaging for assessing brain health include heterogeneous MRI appearances,21 subjective interpretation required for parameters like fractional anisotropy,22 and significant costs, hindering widespread adoption, especially in low- and middle-income countries with limited research funding.
Given the multifaceted and complex nature of brain health, a more holistic approach is necessary. As a result, a comprehensive measure of brain health known as the BHI has been developed as an early endeavor to create such a holistic composite measure. The proposed deep learning models for the computation of the BHI offer a promising avenue for advancing research in brain health development. This incorporates the utilization of three models, wherein one is based on a single-mode approach while the other two adopt multimodal strategies, integrating data from both neuroimaging and assessments. Through the fusion of neuroimaging and assessment data, this strategy facilitates a comprehensive evaluation of brain health, enabling a more nuanced understanding of the impact of interventions on cognitive well-being. Neuroimaging data can enhance the training of models that rely solely on behavioral data. By incorporating neuroimaging data during the training stage, these models can be improved, leading to scalable models that do not require neuroimaging data. The results of this study contribute to the growing body of knowledge in the field of brain health and pave the way for future investigations into personalized interventions for maintaining and enhancing brain health. The main contributions of this study are as follows:
-
The study introduces multiple predictive models that leverage deep learning techniques to predict BHI on a large dataset. Each model is adept at accommodating varying input types, including neuroimaging or assessment data, depending on the available data and the predictive criteria.
-
By employing reconstruction error, the study successfully identifies significant brain regions and assessment data components. This analysis allows researchers to pinpoint specific areas of the brain and key assessment variables that are crucial for our proposed index of brain health.
-
The probability density plots and clustering analysis reveal the presence of distinct subpopulations within the dataset based on BHI. Additionally, the validation of BHI using assessment scores and psychiatric disorder diagnoses further corroborates the reliability of the proposed model.
-
The study investigates the impact of cognition, well-being, lifestyle determinants, and social engagement on brain health. Analyzing these assessment data provides valuable insights into the interplay among these variables and their combined effect on brain health.
-
The research investigates how the brain health index varies across different demographic characteristics. By analyzing how the index changes across different demographic groups, such as age, gender, and socioeconomic status, the study offers valuable insights into the potential influence of these factors on brain health.
This study makes several significant contributions to the field of brain health research. Overall, these contributions advance our understanding of brain health and have practical implications for interventions and personalized approaches in this field.
2. Materials and Methods
2.1. UK Biobank fMRI Data Acquisition and Preprocessing
The neuroimaging training dataset for this analysis was obtained from the UK Biobank database.23 It consisted of 34606 participants, aged 53 to 87 years (mean age: 69.75 ± 7.43 years), including 19120 females (53.1%) and 16880 males (46.8%). The participants underwent rs-fMRI scanning using 3 Tesla (3T) Siemens Skyra scanners with 32-channel head coils. The imaging parameters included a gradient-echo echo planar imaging (GE-EPI) technique with specific settings: no iPAT, fat saturation, a flip angle (FA) of spatial resolution of mm, field-of-view (FOV) of matrix), repeat time (TR) of 0.735s, echo time (TE) of 39 ms, and a total of 490 volumes. The scanning lasted for 6 minutes and 10 seconds, during which participants were instructed to focus on a crosshair and remain relaxed. Eight slices were acquired simultaneously, via a multiband sequence with an acceleration factor of eight.
Various preprocessing procedures were implemented on the UK Biobank database to ensure data quality. To address subject-specific motion, the MCFLIRT tool24 was utilized for intra-modal motion correction. In order to facilitate comparisons of brain scans across participants, grand-mean intensity normalization was applied, scaling the entire 4D dataset using a single multiplicative factor. Residual temporal drifts were mitigated by a high-pass temporal filter, and geometric aberrations were rectified using FSL’s Topup tool.25 EPI unwarping was performed, followed by gradient distortion correction (GDC) unwarping. Independent component analysis (ICA) in conjunction with FMRIB’s ICA-based X-noiseifier26 was employed to eliminate structural artifacts. Furthermore, the data were standardized to an MNI EPI template using FLIRT and SPM12. Finally, Gaussian smoothing with a full width at half maximum (FWHM) of 6mm was applied to the data.
A fully automated spatially constrained ICA process called NeuroMark27 was applied to the rs-fMRI data. We used the Neuromark_fMRI_1.0 template comprising 53 intrinsic connectivity networks (ICNs) that replicated across two large healthy control datasets from a 100-component blind ICA decomposition. These ICNs were then used as templates in an adaptive ICA approach to estimate subject-specific functional networks and their time courses (TCs). Functional network connections were evaluated and categorized into seven domains: subcortical (SC: 5 ICNs), auditory (AUD: 2 ICNs), sensorimotor (SM: 9 ICNs), visual (VIS: 9 ICNs), cognitive control (CC: 17 ICNs), default mode (DM: 7 ICNs), and cerebellar (CB: 4 ICNs). The resulting static functional network connectivity (sFNC) was provided as input to the models that utilized neuroimaging data.
2.2. UK Biobank Brain Health Assessment Data
The assessment data, consisting of self-reported questionnaires, was gathered from 34606 participants in the UKBiobank database. Table 1 shows the different assessment questions and the corresponding brain systems for each question. The assessment questions in the study encompass measures of cognition, mental health, lifestyle factors, and social engagement to evaluate different aspects of brain health. The primary brain system under consideration in the UK Biobank dataset was cognition. It encompasses two essential evaluation parameters: fluid intelligence score and prospective memory. The UK Biobank fluid intelligence test is designed to focus on assessing verbal and numerical reasoning abilities. The test involves participants responding to a series of 13 multiple-choice questions. The computation of the fluid intelligence score involves summing up the correctly answered questions out of the 13 presented within a two-minute duration.
Prospective memory was measured using a single-trial task. Initially, participants were given instructions at the outset of the UK Biobank cognitive test series. These instructions conveyed that they should touch the Orange Circle, instead of the expected Blue Square, when presented with four colored symbols after the tests. Subsequently, participants undertook various other cognitive tests. At the test’s conclusion, participants were shown the four shapes and were prompted to touch the Blue Square. If the participant touched the Orange Circle, signifying the accurate response, the test concluded. Otherwise, if they touched a different shape, a prompt reminded them of the alternative symbol they were supposed to remember and touch. The assigned score in this study was binary: 1 for accurately touching the orange circle initially, and 0 for touching any other shape.
The second brain system employed in computing brain health pertains to well-being. Within this framework, there are 26 evaluation metrics specifically linked to the mental health facet of brain well-being. The 12 assessments from “mood swings” to the “guilty feeling” are specifically designed to derive the neuroticism score of the Eysenck Personality Inventory (EPI-N).28 Neuroticism is a personality trait that encompasses the measurement of emotional stability or instability in individuals. The co-occurrence of neuroticism and an elevated incidence of stressful life events has been found to be significantly associated with a progressive decline in cognitive functioning among elderly individuals who are affected by depression.29 Meanwhile, recent depression symptom (RDS-4) occurrences are summarized in assessments from “frequency of depressed mood in last 2 weeks” to “frequency of tiredness lethargy in last 2 weeks”. It is a continuous measure of symptoms such as sadness, lack of interest, agitation, and fatigue, especially within the past 2 weeks before scanning. The assessments of RDS-4 align with multiple diagnostic criteria outlined in the manual of the Diagnostic and Statistical Manual of Mental Disorders, indicating a possible association with major depressive disorder.30 Also, assessments like “seen a doctor/gp for nerves, anxiety, tension or depression” and "seen a psychiatrist for nerves, anxiety, tension, or depression serve as an indication of the subject’s probable depressive status.31 However, these questions did not distinguish between isolated and recurring depressive episodes. In summary, the set of well-being assessments is completed with the remaining five evaluations, which encompass general satisfaction and the levels of contentment related to family, friendships, health, and financial situation.
Thirdly, the assessment of lifestyle quality encompasses five distinct measures from the UK Biobank database. The first measure focuses on sleep issues, assessing the presence of problems like trouble falling asleep or disruptions during the night.32 Another pivotal aspect is physical activity, which is evaluated through the categorization of activities engaged in over the past four weeks. These activities encompass leisurely walking, vigorous sports, light do-it-yourself (DIY) tasks (like pruning and lawn maintenance), more demanding DIY activities (including landscaping, carpentry, and excavation), as well as other forms of exercise like swimming, cycling, fitness routines, and bowling.33 Alcohol intake is another factor influencing lifestyle, and the evaluation explores an individual’s alcohol-related behaviors. This covers a range from those who completely refrain from alcohol to those who used to drink but have stopped, and to those who currently engage in drinking. Furthermore, dietary patterns are also accounted for in the lifestyle evaluation. Variations in the diet on a week-to-week basis, as well as any significant alterations to dietary habits within the past five years, contribute to the comprehensive assessment of one’s lifestyle quality.
The last part of the assessment focused on the brain system related to social life, involving two evaluations. The initial assessment determined the frequency of engagement in various social leisure activities, such as going to the gym, participating in social clubs, religious groups, adult education classes, and other group activities.34 The second evaluation gauged the regularity of visits to friends or family, offering response options ranging from “almost daily” and “2-4 times a week” to “never or almost never,” providing insight into participants’ social interactions. The selection of these assessment variables was driven by a combination of factors, including the multidimensional concept of brain health,35 the availability of measures within the UK Biobank dataset, and prior research utilizing similar measures for mental health and cognitive decline studies.30,36 Numerous affect-based mental health measures are available in the UK Biobank dataset. Neuroticism was evaluated using the 12-item Eysenck Personality Questionnaire-Revised Short Form (EPQ-RS),37 corresponding to the initial 12 assessments in the well-being section. Higher neuroticism scores indicate heightened susceptibility to negative emotions like anxiety, worry, fear, anger, frustration, and loneliness. Inquiries 16-19 in the well-being section focus on recent depressive symptoms (RDS-4), a continuous measure recorded during scanning, assessing feelings of low mood, indifference, restlessness, and weariness. Additionally, Smith and his colleagues introduced a categorical measure of lifetime depression incidence using questions 14 and 15, indicating potential depressive status.31 Meanwhile, within the cognitive assessment, integrated into the fully-automated touchscreen questionnaire, prospective memory and verbal and numerical reasoning (Fluid Intelligence) were evaluated.38 Additionally, recent studies have identified robust associations between sleep and mental health (Hepsomali and Groeger 2021b), along with diet and cognitive measures (Hepsomali and Groeger 2021a) within the UK Biobank dataset.
2.3. Methods
2.3.1. Variational Autoencoders
The variational autoencoder39 is an unsupervised generative deep learning model that offers a probabilistic framework for characterizing observations in latent space while simultaneously generating new samples. The architecture of the VAE mainly consists of an encoder and a decoder. Unlike traditional autoencoders that produce a single value to represent each latent attribute on the encoder side, the VAE uses probability distributions for describing observations in the latent space. In a VAE, the encoder network transforms the input data to a latent space, typically represented by a multivariate Gaussian distribution. This transformation is characterized by two sets of parameters: the mean and the variance which define the distribution in the latent space. These parameters are then used to sample a latent vector that is representative of the input data. The sampling process is obtained by reparametrizing the latent vector as follows:
\[z=\mu+\sigma\odot\epsilon\tag{1}\]
where is a random variable sampled from a standard Gaussian distribution The decoder network then takes this latent vector and maps it back to the original data space, aiming to reconstruct the input. During training, the VAE optimizes a loss function that minimizes both the reconstruction loss and the Kullback-Leibler (KL) divergence between the distributions of the latent variables and independent normal distributions.40 This reconstruction error essentially measures the difference between the original input and the reconstructed output.
\[Reconstruction\hspace{0.1 in} Error=\frac{1}{N}\sum_{i=1}^N (x_{i}-\hat{x_{i}})^2\tag{2}\]
where and N denote the original input, reconstructed output, and number of samples, respectively.
2.4. Brain health index prediction framework
This study presents a novel approach to predicting the brain health index using three distinct models. Each model serves a unique purpose by leveraging different types of data for both training and testing. The first model relies exclusively on assessment measures during its training and testing phases. The second model takes advantage of both neuroimaging and assessment data during its training, but during testing, it only utilizes assessment data. This highlights the potential of incorporating neuroimaging data to enhance model training while still being able to make predictions when only assessment data is available. The third model, encompassing the full scope, employs both neuroimaging and assessment data for both training and testing. This showcases the comprehensive approach of utilizing all available data modalities for accurate brain health index prediction.
The process of predicting BHI in the three distinct cases involves two primary stages. In the initial stage, PCA feature extraction is utilized to decrease the complexity of the dataset’s information while preserving the most essential features. In the subsequent phase, the significant features obtained from the initial PCA step are employed to create a feature vector. For instance, in the second and third models where both neuroimaging and assessment data are utilized for training, the feature vector is formed by concatenating the dimensionality-reduced assessment and neuroimaging features. This feature vector is then used as input for a VAE, which generates a compact representation of the features. This approach aims to enhance the prediction of BHI by effectively capturing the pertinent patterns within the data.
In general, the VAE consists of encoder and decoder components that work in conjunction to process the input data. In this proposed network, the encoder, consisting of four fully connected hidden layers, progressively reduces the dimensionality of the data. The first hidden layer contains 16 nodes, followed by layers with 8, 4, and 2 nodes, respectively. This encoder network produces a compressed representation of the input data in the latent space. Conversely, the decoder exhibits a symmetric structure with the encoder, which is also composed of four hidden layers. The first hidden layer contains 2 nodes, followed by layers with 4, 8, and 16 nodes. The decoder’s output layer aims to reconstruct the original input data. The selection of node configurations in the encoder and decoder architecture underwent a rigorous cross-validation process aimed at assessing and optimizing their efficacy. Multiple configurations were systematically tested and compared, considering their impact on the model’s performance metrics, such as reconstruction accuracy and generalization ability.
Here, the use of both PCA and a VAE in conjunction for dimensionality reduction serves distinct yet complementary purposes. PCA is initially employed as a linear technique to condense the data by emphasizing the most substantial variance while preserving essential features, although potentially missing intricate non-linear associations within the dataset. In contrast, the VAE, being a non-linear method, has the potential to capture these nuanced and complex patterns that PCA might overlook. Rather than just compressing the data, the VAE learns to encode and decode the information, aiming to reconstruct the input accurately. This process of encoding and decoding results in a representation that not only reduces dimensions but also captures more intricate and detailed patterns in the dataset. These patterns may include non-linear associations and dependencies between features.
During training, the VAE minimizes the reconstruction error, which quantifies the disparity between the original input and the reconstructed output. This is accomplished through a combination of reconstruction loss and KL loss, constituting the VAE loss. In this study, the Adam optimizer41 was used to train the VAE for 1000 iterations, with a learning rate of 0.001 and a batch size of 32. After training the VAE, the output includes a latent variable along with its corresponding mean and variance Through experimentation with varying dimensionalities, we systematically evaluated the performance of the VAE in terms of reconstruction error. The chosen dimensionality for the latent variable is 2, resulting in two distinct variables, namely z1 and z2. We explored a range of dimensions, from lower values such as 1 to higher ones like 10 or more. It was observed that as we increased the dimensionality beyond 2, the reconstruction error either plateaued or exhibited marginal improvements, which did not justify the added complexity associated with higher-dimensional latent spaces. Using the two latent variables, the BHI can be calculated using the following formula:
\[BHI= z1\times \sigma1+z2\times \sigma2\tag{3}\]
Fig. 1 depicts the architecture of the proposed model, providing a detailed exposition of the feature extraction and BHI prediction processes for each of the three distinct cases.
3. Experimental Setup and Results
To assess the efficacy of the proposed model, systematic experiments were conducted on three complementary models for the BHI prediction. The dataset employed for this purpose encompassed 34,606 participants sourced from the UK Biobank database. Within this dataset, 60% of the participants were allocated for training, while 20% each were dedicated to validation and testing. Various cases were analyzed to calculate the BHI by employing different data modalities for training and testing. A detailed description of these cases is provided below:
-
Case 1: Calculation of BHI using assessment data for both training and testing.
This model relies exclusively on the 34 assessment measures obtained from the UK Biobank dataset to predict BHI. Within the training phase, these assessment measures are passed through the feature extraction block to extract the most significant features. This block employs PCA on the assessment features for dimensionality reduction. Subsequently, the data is fed into the VAE stage. Within the VAE, the data is processed to learn the underlying distribution of the feature vectors, ultimately producing a compact, lower-dimensional representation of the assessment data. The training and validation of this model continue until convergence. Subsequently, the fine-tuned VAE model is evaluated using the test data, which consists solely of dimensionality-reduced assessment measures. The VAE generates a meaningful, low-dimensional representation of the test data, which is then employed to estimate the BHI. To assess the model’s performance, the entire experiment is repeated after excluding the feature extraction block. This allows for a direct comparison of the outcomes from the two scenarios. -
Case 2: Calculation of BHI by training with both assessment and sFNC data but testing using solely assessment data.
This model employs a combination of neuroimaging and assessment data for its training process. It focuses on utilizing the upper triangular segment of the sFNC matrix, resulting in the utilization of 1378 features from the sFNC data and 34 features from the assessment data during the training phase. In the training stage, a parallel PCA feature extraction step is executed to lower the dimensionality of both the neuroimaging and assessment data. The resultant feature vectors from these two data types are concatenated and then fed into a VAE. The primary role of the VAE is to capture the inherent distribution within these feature vectors, ultimately generating a compressed representation of the input data possessing fewer dimensions. The training and validation processes for this model are conducted until convergence is reached. Following this, the fine-tuned VAE model undergoes evaluation using test data, which exclusively comprises dimensionality-reduced assessment measures. The condensed representation produced by the optimized VAE model is subsequently used to estimate the BHI. -
Case 3: Calculation of BHI by training and testing with both assessment and sFNC data.
This model uses both neuroimaging and assessment data for training and testing. The training phase is like case 2, where the PCA method simplifies the neuroimaging and assessment data. These dimensionality-reduced feature vectors are combined and then fed into the VAE. However, during testing, the VAE model is assessed using dimensionality-reduced assessment measures and sFNC data. Additionally, this experiment is also repeated without the feature extraction block.
Fig. 2 provides a probability density plot to visualize the distribution of the BHI for case 1 for 6922 test subjects. In this case, the probability density plot of BHI derived solely from assessment data reveals a range spanning from -3 to 4. The presence of two distinct Gaussian peaks suggests that there are two predominant states within the data. These peaks may correspond to different subpopulations within the dataset, each exhibiting a characteristic brain health level. In the second case, where both neuroimaging and assessment data are employed for training, while only assessment data is used for testing, the BHI range is extended from -4 to 4. The probability density plot now exhibits three Gaussian peaks as shown in Fig. 3. The central peak, being the tallest among the three, suggests that a significant portion of the dataset exhibits a relatively moderate brain health index. The presence of additional peaks on either side of the central peak signifies two distinct subgroups, possibly representing individuals with higher and lower brain health indexes. The use of sFNC data during training has allowed the model to capture more complex patterns, leading to the emergence of a third peak. Finally, in the third case, where both neuroimaging and assessment features are utilized for both training and testing, the BHI range spans from -6 to 6. Similar to the second case, the probability density plot in Fig. 4 presents three Gaussian peaks. The central peak, once again the most prominent, signifies a dominant brain health level within the dataset. The presence of the same number of peaks in cases 2 and 3 indicates that even with the extended BHI range, the underlying distribution remains relatively stable. The richer feature set from the combined data sources might have facilitated the increased BHI range, leading to broader distribution while still maintaining the characteristic central peak representing the most prevalent brain health state.
3.1. Identification of subpopulations and validation of BHI
In all three cases, the presence of Gaussian peaks indicates the existence of underlying subpopulations within the dataset, each characterized by varying brain health indexes. The differences in the number of peaks, their heights, and the BHI ranges highlight the influence of the data sources and training methodologies on the resulting probability density plots. This interplay between data types and training strategies provides insights into the complexity of brain health assessment and the potential benefits of incorporating multiple data modalities in the analysis. Additionally, the presence of different subpopulations was verified by conducting a clustering analysis as shown in Fig. 5. For case 1, the optimal number of clusters was found to be 2, while for cases 2 and 3, the optimal clusters were found to be 3. This further underscores the presence of distinct subpopulations within the dataset, reinforcing the significance of the observed Gaussian peaks.
To validate the accuracy of the BHI, violin plots were generated to compare BHI values across different categories of assessment scores. Each subject’s assessment score resulted from summing responses across all the assessment measures provided in Table 1. By employing k-means clustering, it was observed that in case 1, subjects fell into only two categories with low and high assessment scores. Conversely, cases 2 and 3 revealed three distinct categories corresponding to low, medium, and high assessment scores. Subsequently, BHI values were extracted for each group, and the mean and standard deviation were calculated for every category. The resulting violin plot in Fig. 6 offers insights into the correlation between assessment scores and BHI levels. Before the analysis, data preprocessing involved normalizing both assessment scores and BHI values to a range of 0 to 1. In case 1, subjects with low assessment scores displayed a mean BHI value of 0.45, whereas those with high assessment scores showcased a notably higher mean BHI value of 0.69. Conversely, in case 2, the distinction was more pronounced: subjects with low assessment scores had a mean BHI of 0.39, medium scorers had a mean BHI of 0.45, and high scorers had the highest mean BHI of 0.71. Similarly, in case 3, a comparable trend was observed, although the differentiation between low and medium assessment scores was subtle. These statistical differences were further validated using independent sample t-tests. In case 1, the p-value between low and high assessment scores was less than 0.05, revealing a statistically significant difference between the two groups. Conversely, in case 2, all three categories exhibited p-values below 0.05, signifying statistically meaningful distinctions within each category. However, in case 3, the p-value comparing the low and medium assessment groups exceeded 0.05, suggesting no statistically significant difference between them.
To broaden the validation of BHI across diverse clinical subgroups, two primary categories were selected: individuals with psychiatric disorders and those with cardiovascular diseases. Psychiatric disorders exert distinct yet substantial effects on multiple facets of brain structure and function, making them significant for assessing the effectiveness of the proposed BHI. Within these disorders, particular attention was given to bipolar disorder and major depression due to their high impact on mental health and cognitive capabilities. Fig. 7 displays a violin plot comparing the mean and standard deviation of BHI among individuals diagnosed with bipolar disorder and major depression disorder using data from the UK Biobank dataset. Across all three cases examined, it is evident that the BHI values are consistently low, with means below 0.5. Compared with bipolar disorder, subjects with major depression disorder have a slightly higher mean BHI. This difference suggests that within this dataset, subjects with bipolar disorder tend to exhibit poorer brain health than those diagnosed with major depression disorder. Moreover, with all cases yielding p-values below 0.05, it is evident that there exists a statistical distinction between these groups to BHI.
Subjects with cardiovascular diseases, such as heart attack, stroke, and high blood pressure, were included due to their significant impact on brain health. Fig. 8 shows a violin plot comparing the mean and standard deviation of BHI among individuals with these conditions. In all cases, the subgroups have a mean BHI of less than 0.5. Cases 1 and 2 reveal statistically significant differences among all three groups, while Case 3 shows no statistically significant difference between heart attack and high blood pressure categories. This indicates that while heart attack and high blood pressure both negatively affect brain health, their impacts may not be distinguishable in this subgroup using the method applied in Case 3. Therefore including these cardiovascular conditions allows for a comprehensive assessment of their specific and combined effects on the proposed BHI.
Table 2 presents the results of evaluating the reconstruction error for predicting BHI using only assessment data. The table highlights the impact of applying PCA dimensionality reduction on the reconstruction error. Evidently, the magnitude of the reconstruction error seems to fluctuate in accordance with the number of assessment features utilized for prediction. Notably, opting for the scenario where PCA dimensionality reduction is omitted and instead utilizing all 34 assessment features resulted in the lowest reconstruction error (0.0805 for training and 0.0809 for testing). This result indicates that the given configuration offers the highest level of accuracy when estimating the BHI while utilizing the assessment data. Also, the model demonstrates effective generalization as the differences between training and testing errors are relatively small.
Table 3 depicts the evaluation of the reconstruction error for BHI prediction for case 2. The table explores various scenarios, each characterized by different combinations of PCA dimensionality reduction for sFNC and assessment data, as well as different numbers of sFNC and assessment features used in training. In instances where PCA was applied, the number of sFNC/assessment features in the table indicates the number of principal components after dimensionality reduction. The best BHI prediction performance is achieved when the training dataset includes a balanced combination of sFNC and assessment features. The scenario utilizing 10 FNC features and 10 assessment features demonstrates the lowest reconstruction error, highlighting the importance of considering both types of data for accurate predictions. Notably, the experiment that excludes dimensionality reduction couldn’t be tested for case 2 due to the potential mismatch in dimensionality that could arise during testing. This mismatch stems from exclusively utilizing assessment features during testing, while the training phase involves both sFNC and assessment features.
A comprehensive evaluation of reconstruction errors related to case 3 is presented in Table 4. In this scenario, BHI prediction is conducted using a fusion of sFNC and assessment data for both the training and testing phases. The study explores various combinations of dimensionality reduction techniques, feature sets, and feature numbers to understand their impact on the quality of BHI prediction. The primary emphasis lies in assessing reconstruction errors, encompassing both the training and testing stages, with distinct values presented for models based on sFNC and assessment data. The most significant result is employing PCA-based dimensionality reduction on both FNC and assessment data. This involved reducing sFNC features to 1000 dimensions and assessment features to 30 dimensions. This configuration achieved a low reconstruction error of 0.0681 for training and 0.0667 for testing. This indicates that the chosen combination of data sources, feature reduction, and feature count led to a model that effectively captures and reproduces the underlying patterns in the data.
3.2. Variation in BHI with age, gender, and education
This analysis focuses on examining the distribution of BHI in relation to demographic characteristics such as age, gender, and educational qualification. The test study encompassed a total of 6922 participants, whose ages ranged from 53 to 86 years. The median age of the participants was 70 years, and the interquartile range (IQR) of their ages spanned from 64 to 75 years. In terms of gender distribution, the sample consisted of nearly equal proportions of males (46.6%) and females (53.4%), ensuring a balanced representation of both sexes in the analysis. Regarding the participants’ educational backgrounds, the study group was notably well-educated. Specifically, 45.7% of the participants held a college or university degree, highlighting a substantial proportion of higher education attainment. Additionally, 12.9% possessed a higher school certificate equivalent to A levels/AS levels, typically achieved around age 18. Also, 19.1% had a certificate similar to O levels/GCSEs, attained after 10 years of school, around age 16. A smaller percentage, 3.9%, had a certificate of secondary education (CSE) or an equivalent qualification, also acquired after 10 years of school, like O Levels/GCSEs. Furthermore, 5.6% of participants had obtained a higher national certificate or diploma, such as NVQ, HND, or HNC, which come after secondary school and usually take 1-2 more years. A category labeled ‘other professional qualifications’ was represented by 4.7% of the sample. Notably, 8.2% of participants did not fall into any of the educational categories. This category represents individuals who might not have completed formal education up to the levels mentioned above.
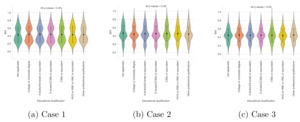
In this analysis, a violin plot was utilized to visually portray the distribution and essential statistical metrics of BHI across distinct categories, such as age, gender, and education. Additionally, independent samples t-tests were conducted to compare the BHI across different demographic groups. The BHI is normalized to be between 0 and 1, with a mean value of 0.5. Fig. 9 specifically presents the plot between educational qualifications and BHI for cases 1, 2, and 3, along with the corresponding p-values. The elongated violin plots comprise seven distinct educational levels. The results from the t-test indicated no statistically significant differences between the groups, as all p-values were above 0.05. This type of distribution indicates that there is no strong bias or trend in one direction of BHI. Subsequently, Fig. 10 illustrates the relationship between gender and BHI for the different cases. Notably, both male and female groups exhibit analogous features with violins of varying widths and heights. This similarity signifies comparable levels of BHI variation across genders. With p-values of 0.116, 0.503, and 0.174 for cases 1, 2, and 3, there is no statistical significance in BHI between females and males. The third graphical representation, depicted in Fig. 11, displays the relationship between BHI and age. Notably, this plot bears a resemblance to the trend observed in the gender violin plot. Overall, these plots highlight the variability in BHI scores across different categories of education, gender, and age. In all three plots, the t-test with p-values greater than 0.05 implies that the demographic categories under consideration might not have significantly influenced the BHI scores.
3.3. Identifying significant contributors to BHI: brain regions and assessment measures
In VAE, the most significant features that contributed to the performance of the model can be found by computing the reconstruction error. The reconstruction error is used as a metric to assess the quality of the generated data and evaluate the performance of the model. The VAE learns to identify and represent the prominent features of the input data in the latent space by minimizing the reconstruction error.
Measuring feature importance using reconstruction error in variational autoencoders involves assessing the impact of each input feature on the model’s ability to accurately reconstruct the original data. In the context of the three testing cases, the evaluation involves calculating the reconstruction error using the VAE model. The first case focuses solely on utilizing assessment measures from the UK Biobank database as input to the VAE. The resulting plot (Fig. 12) illustrates the reconstruction errors of the VAE using the 34 assessment features. Notably, the highest reconstruction error observed in this case is 0.21. Moving to the second case, the VAE is tested using only the assessment measures, even though its training encompassed both sFNC and assessment data. The corresponding reconstruction error plot (Fig. 13) demonstrates the performance of the VAE using the same 34 assessment features. Within this plot, the most significant reconstruction error recorded is 0.23. In the third case, the VAE’s assessment involves inputs from both sFNC and assessment measures. The evaluation results in two distinct plots: Fig. 14, showcasing the reconstruction errors using the 34 assessment features, and Fig. 15, depicting the sFNC domains with the highest reconstruction errors. In the assessment-focused plot, the highest reconstruction error observed is 0.25.
In the case of multimodal inputs, the VAE aims to capture the inherent structure and characteristics of each modality in the latent space and subsequently reconstruct them. However, different modalities might exhibit varying degrees of complexity or information content, leading to differences in reconstruction errors. For instance, sFNC data contains more detailed and distinctive features compared to assessment data. Hence, the reconstruction error for that modality may be higher, indicating a greater challenge in effectively reconstructing it. Conversely, assessment data contains less complex information and thus has a lower reconstruction error.
During the testing phase for case 1, the assessment data revealed three prominent features with the least reconstruction error: 1) family relationship satisfaction; 2) illness, injury, or bereavement stress in the last 2 years; and 3) risk-taking. Similarly, during the testing phase for cases 2 and 3, the assessment features with the least reconstruction error were family relationship satisfaction, illness, injury, bereavement stress in the last 2 years, and work-job satisfaction. The consistent emergence of these patterns across all three cases suggests that the computation of the BHI is significantly influenced by these shared attributes within the assessment data. In the case of the sFNC matrix for case 3 testing, the SM, VS, and CB domains showed the highest reconstruction error.
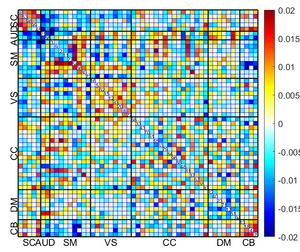
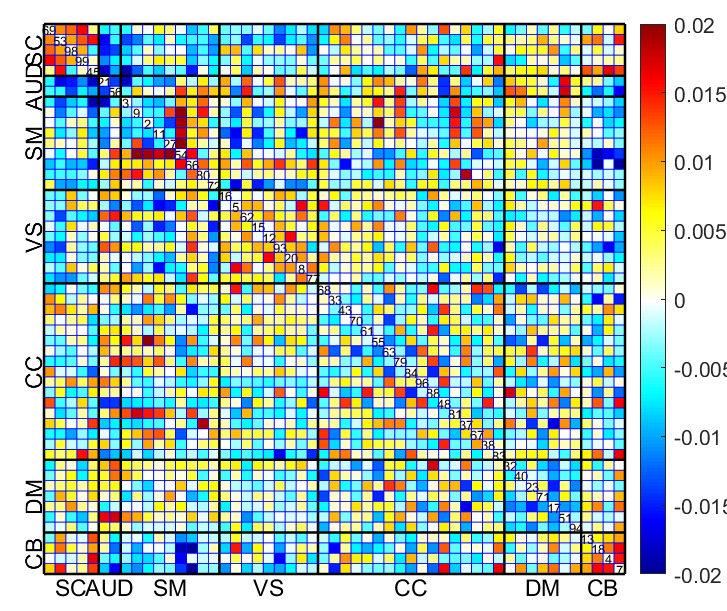
Fig. 16 displays the mean sFNC matrix of 25% of subjects with the lowest BHI among the total of 6922 test subjects. Conversely, Fig. 17 shows the mean sFNC matrix of 25% of subjects with the highest BHI among the same group of 6922 subjects for the same time points. Significantly, individuals displaying the highest BHI values exhibited enhanced connectivity in the SC and select regions of the SM. Conversely, participants with the lowest BHI values demonstrated heightened connectivity in the SM, VS, and SM-VS domain pairs. These observations obtained from the mean sFNC data of the UK Biobank study align with the regions identified through the calculation of reconstruction error.
The bar graphs presented in Fig. 18 depict the reconstruction error concerning distinct brain systems across multiple scenarios. It can be noted that, for case 1, the lifestyle assessment exhibits the highest reconstruction error. This suggests that the model struggles to accurately reconstruct or predict the lifestyle assessment measure based solely on the assessment data. Conversely, for cases 2 and 3, the well-being metric demonstrates the highest reconstruction error. This suggests that even when both assessment and neuroimaging data are used for training and testing, the model still struggles to predict the well-being assessment measure effectively.
4. Discussion
In this work, we propose three novel complementary approaches for BHI prediction using deep learning. Developing a practical approach for quantifying brain health is critical in understanding the impact of interventions aimed at enhancing cognitive well-being and mitigating neurodegenerative diseases. This research tackles this pivotal challenge by introducing both unimodal and multimodal systems that facilitate efficient BHI computation. By leveraging a large dataset, the deep learning model captures all the variations of brain connectivity, providing a comprehensive assessment of brain health. Our approach aligns with previous studies that have utilized deep learning techniques for neuroimaging data analysis.42,43 Specifically, the use of sFNC data also serves as a valuable neuroimaging feature to draw inferences on early cognitive and psychiatric behaviors in both adults and children.44 Moreover, the incorporation of multimodal data integration with neuroimaging data is supported by literature suggesting that it can enhance the accuracy of mental disorder predictions.45–47
Table 1 details the assessment measures from the UK Biobank dataset used in our study. The psychometric properties of the assessment measures are crucial for ensuring the reliability and validity of the BHI. Encompassing various domains such as cognition, well-being, lifestyle, and social life, they provide a comprehensive evaluation. For instance, in related works, the fluid intelligence score has demonstrated high reliability and validity in measuring cognitive function.48 Similarly, assessments related to mood and mental health, such as those capturing irritability, anxiety, and depression symptoms, have been validated in large cohorts and shown to possess reliable psychometric properties.49 Consequently, these assessment measures have been utilized individually in numerous studies focusing on mental health and various psychiatric disorders. However, these measures have not been combined to create a comprehensive BHI, which underscores the novelty of our approach.
The analysis is trifold, with the initial case employing solely unimodal input with assessment data. The subsequent two scenarios, however, leverage multimodal input by synergizing neuroimaging and assessment information. Even though analysis conducted using just the assessment data produced meaningful outcomes, the utilization of rs-fMRI data provides valuable insights into the intrinsic functional organization of the brain during a non-task state. To extract significant features from the neuroimaging and assessment data, the model employs dimensionality reduction using PCA. Later, by incorporating VAE in the subsequent stage, the model learns a low-dimensional representation of the input data that captures its essential characteristics. This compressed representation facilitates the calculation of the BHI, enabling a more concise and interpretable assessment of brain health. Following this, the assessment of the BHI was done during both training and testing for all three cases. The results revealed that in the first case, the configuration devoid of PCA demonstrated superior performance by achieving the least reconstruction error. In cases 2 and 3, PCA dimensionality reduction was required for both the sFNC and the assessment data to achieve the least error. Additionally, sFNC data exhibited a lower reconstruction error in comparison to assessment data. This observation underscores the VAE’s capacity to model the underlying patterns present in the neuroimaging data, thereby enabling it to reconstruct the sFNCs with greater accuracy and detail. The study also examined how variations in BHI relate to demographic characteristics such as age, gender, and education. Across different demographic attributes, t-tests revealed no statistically significant differences in BHI between the groups. This underscores the comprehensive and robust nature of the BHI, which integrates multiple aspects of brain function and structure. As a result, the BHI is less susceptible to demographic variation, providing consistent results across different populations.
The proposed approaches were evaluated for interpretability by identifying significant assessment measures and sFNC domains. Elevated reconstruction errors were observed in specific regions within the SM, VS, and CB domains, indicating inaccuracies in capturing connectivity patterns. BHI, based on four brain systems (well-being, cognition, lifestyle, and social life), showed varying reconstruction errors across cases. Social life and cognition consistently had the lowest errors, while well-being had the highest in cases 2 and 3. This difficulty arises from the inherent complexity of well-being as it encompasses a wide range of subjective experiences, emotions, and personal perceptions. Meanwhile, relying solely on assessment data for well-being helped to improve prediction accuracy by reducing noise and complexity introduced by neuroimaging data. This study also identified the presence of Gaussian peaks in the probability density plots of the BHI across three distinct cases suggesting the existence of underlying subpopulations within the datasets, highlighting the heterogeneity of brain health profiles. Clustering analysis further validates this, revealing distinct subgroups within the data. The correlation between assessment scores and BHI underscores the importance of comprehensive evaluation approaches. While in case 1, there is a clear distinction between subjects with low and high assessment scores with corresponding BHI levels, cases 2 and 3 demonstrate a more complex relationship, with multiple assessment score categories reflecting varying BHI distributions.
Furthermore, the comparison of BHI between subjects with bipolar disorder and major depressive disorder from the UK Biobank dataset also yielded significant insights. While both conditions exhibit lower BHI values, there is evident variability within each disorder cohort. This variability underscores the heterogeneous nature of psychiatric disorders and highlights the diverse impacts they may exert on brain health across individuals. Cognitive dysfunction is a recognized feature of these mood disorders, including major depressive disorder and bipolar disorder.50,51 The observed differences in BHI between individuals with bipolar disorder and major depression disorder align with existing knowledge suggesting that cognitive impairment is more pronounced in bipolar disorder. Meanwhile, in the case of cardiovascular disease, prior neuroimaging studies have demonstrated that individuals with a higher risk of these diseases have structural and functional brain alterations.52,53 Therefore, our study also focused on individuals with a history of heart attack, stroke, and high blood pressure. Statistical analysis (p-values) in cases 1 and 2 showed significant differences among all three groups, suggesting varying impacts on brain health among these conditions. However, while heart attack and high blood pressure both negatively affect brain health, their impacts were not distinguishable using certain assessment methods, as seen in Case 3. Overall, the findings from our analysis offer valuable insights into the complexities of brain health assessment and the advantages of employing diverse data modalities and assessment methodologies.
5. Conclusion and Future Works
This study introduced three innovative and complementary approaches for predicting BHI using deep learning techniques. By integrating both unimodal and multimodal strategies, this research presents novel approaches to computing BHI that have not been explored in previous studies. The analysis progresses through three phases, starting with a unimodal strategy that utilizes assessment data and then evolving into multimodal configurations that combine neuroimaging and assessments. The application of dimensionality reduction methods like PCA during data preprocessing facilitated the extraction of essential features necessary for BHI computation. The subsequent integration of VAE enabled a compact representation of input data, enhancing the precision of BHI calculations. Throughout training and testing, all three cases underwent BHI assessment, comparing various dimensionality-reduced features to identify optimal configurations with minimal reconstruction error. In the case that relied solely on assessment data, the exclusion of PCA resulted in the lowest reconstruction error, underscoring the unique requirements of each case. Multimodal setups exhibited superior reconstruction accuracy, showcasing the model’s adeptness at capturing neuroimaging patterns.
Moreover, the identification of distinct subpopulations within the dataset based on BHI highlights the heterogeneity of brain health profiles, influenced by data sources and training methodologies. Validation through assessment scores reveals correlations between assessment measures and brain health levels, with multimodal approaches showing enhanced predictive power. The lower BHI values among individuals with psychiatric disorders and cardiovascular diseases reveal insights into the impact of these conditions on brain health. Additionally, demographic attributes such as age, gender, and education did not exert a notable influence on BHI across the different cases. The interpretability of the approach was assessed by identifying significant assessment measures and sFNC domains, emphasizing the relevance of different brain systems and regions. Overall, this work contributes to the development of a practical approach for quantifying brain health, which is crucial for understanding interventions aimed at enhancing cognitive well-being and mitigating neurodegenerative diseases.
However, this study has its limitations, with one notable constraint related to the sFNC measures. While sFNC offers valuable insights into the brain’s functional organization, it may overlook the dynamic nature of brain connectivity. Static measures might miss transient but significant changes in brain activity, potentially compromising the accuracy of the BHI. Meanwhile, assessment measures, even though comprehensive, are susceptible to biases and inaccuracies arising from self-reporting and subjective interpretation. These limitations in both sFNC and assessment measures mildly impede the potential use of the proposed BHI by introducing noise and reducing the reliability of the computed index. Furthermore, the current findings stem from a single-center study, emphasizing the need for broader validation. Hence, future research includes validating these methodologies on an independent dataset to reinforce and extend the reliability and applicability of our approaches across diverse settings and populations. Future studies will also explore the use of dynamic functional connectivity measures, more objective assessment metrics, and cross-validation of self-reported data with clinical evaluations. Additionally, we plan to investigate the integration of attention-based models, such as transformers, to further enhance the overall predictive performance of our deep learning approach in forecasting BHI. Leveraging insights from this multimodal framework, we aim to develop personalized interventions tailored to individuals based on different brain systems. By harnessing the power of deep learning, we envision creating targeted interventions that effectively promote and optimize brain health in diverse populations.
ACKNOWLEDGEMENTS
This work was supported by the Georgia State University RISE program, NSF grant 2316421 and NSF grant 2112455.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflict of interest.
DATA AVAILABILITY STATEMENT
All data is publicly available at the UK Biobank online database https://www.ukbiobank.ac.uk/.