Introduction
In the last decade, data sharing in neuroimaging has been increasingly encouraged in order to improve reproducibility and translation of research findings into clinical use. Openly shared datasets, ranging from grassroot resources such as the 1000 Functional Connectome Project1 or the Autism Brain Imaging Data Exchange (ABIDE2), to resources directly funded for the purpose of being shared with the community, such as UK Biobank3 or the Erasmus Glioma Database,4 have created new opportunities and changed the neuroimaging research landscape. In parallel with those large-scale initiatives, complementary efforts have aimed to facilitate sharing of smaller datasets (so called “long tail of science”5). These efforts have notably led to the development of standards for data organization (such as the Brain Imaging Data Structure (BIDS)6), but also to new data sharing platforms, such as OpenNeuro,7 or earlier, the fMRIDC8 in neuroimaging as well as generalist repositories.9
On the benefits of data sharing
Data sharing benefits neuroimaging research by boosting statistical power, enabling new types of analyses, enhancing population representation, and improving reproducibility. First, collaboration and pooling of data across acquisition sites is a means to increase the sample sizes of research studies. Combined with other approaches such as optimizing the design of the experiment,10 increasing sample size can be an important lever to achieve sufficient statistical power. The lack of statistical power of neuroscience studies has long been criticized,11,12 in particular for its impact on false positive findings. In addition, pooling data is also a requirement in the case of rare diseases research, such as rare brain tumors,13 for which the possibility to recruit participants from a single site is inherently limited. Second, with the development of analysis tools based on machine learning and deep learning techniques, larger sets of data (that may be impossible to obtain in a single center) are typically required. This is important for several applications relating to image biomarker development and validation, or radiomics. Third, data sharing enables the creation of more representative datasets that better reflect the general population. An ample literature (see for example14 in brain imaging or15 and16 from preclinical research) has demonstrated how diversity in research data is a requirement to avoid biased classifiers. Finally, a growing momentum drives the sharing of individual datasets in support of research findings.17–19 This is a key milestone for computational reproducibility (i.e., ensuring that identical results can be reproduced under the same conditions, including data, code, and methodology).
Beyond improving reproducibility and trust in scientific results, building shared datasets also creates opportunities to shape and transform the scientific culture and how science is conducted. Shared datasets provide educational opportunities and foster methodological innovation. In addition, they support global collaboration and cross-disciplinary research, enhancing the integration of interdisciplinarity into medical research. Data sharing unlocks the potential of existing datasets to generate new research ideas and hypotheses and address different research questions using the same data,20 thus ensuring a more efficient use of the participants’ time,21 scientific funding and resources.
Support from funding bodies and governments
The sharing of research data is increasingly supported by funding bodies and governmental agencies, which recognize its critical role in advancing scientific research. Funding organizations such as the European Research Council (ERC) and the National Institutes of Health (NIH) often require grant recipients to adhere to open data policies.22,23 Across Europe, many countries have implemented national frameworks and incentives to foster open science. These are very heterogeneous and range all the way from simple open access up to mandatory data sharing. Examples of European bodies with open science policy include the European Union (EU)'s and its Horizon Europe program,24 the French National Plan for Open Science,25 the Belgium FWO[1], the Dutch Research Council (NWO)[2], the Swiss National Science Foundation[3], Portugal’s Foundation for Science and Technology[4] and the German Research Foundation[5].
Best practices for data sharing
Throughout the years, best practices have emerged from the scientific community, in particular, shared data should follow the FAIR and CARE guiding principles. According to FAIR, data should be Findable (i.e., data should be easy to locate), Accessible (i.e., data must be available to users), Interoperable (i.e., data should be formatted and structured in a way that allows it to work with other datasets and systems), and Reusable (i.e., data must be well-documented and licensed for use by others).26 The CARE principles state that data should have a Collective benefit (i.e., data sharing should prioritize the benefits to communities and stakeholders involved), Authority (i.e., this principle emphasizes the right of individuals and communities to govern their own data), Responsibility (i.e., those sharing and using data must take accountability for their actions), and Ethics (i.e., the process must be conducted with respect, transparency, and fairness, particularly regarding sensitive or personal data).27,28 By integrating the FAIR and CARE principles, data sharing can be more effective and responsible, ultimately leading to greater trust and collaboration in research and data use.
Additionally, incorporating patient and public perspectives is key to overcoming these challenges. Privacy loss, surveillance, and discrimination risks erode trust and reduce willingness to share data. To address this, frameworks must reflect societal values — ensuring fair benefit distribution, accountability, and open dialogue. Public involvement can be achieved through deliberative polls, citizen juries, participatory appraisals, scenario-based workshops, focus groups, and participant-led data cooperatives, as well as direct roles in data governance. For instance, dynamic consent platforms let users adjust their preferences over time.29,30 Ultimately, these participatory approaches enhance transparency, accountability, and public trust in science.
Data sharing and privacy
Compliance with data protection laws, such as the General Data Protection Regulation (GDPR) in the EU (see Appendix 1 for more details) but also with equivalent regulations in non-EU countries (see Appendix 2 for an illustration), has provided important safeguards for privacy and security in neuroscience research. This legislation refers to the fundamental right of human participants to have their identity and privacy protected and has been in effect for all processing of personal data of EU citizens since 2018[6]. While these regulations aim to enhance data protection and privacy, they also introduce significant requirements in data management and sharing practices. Data protection laws vary across countries,31–33 creating ambiguity for researchers, who may struggle to determine which platforms they are legally permitted to use.
An example of variability in definitions across geographical locations is the difference between the notion of anonymization in the United States (US) and the EU’s GDPR.34 In the US, the definition emphasizes primarily identifiable features (i.e., name, birth date, face, etc.) while the European definition states that if data can be individualized to a single participant (i.e., if data is a unique signature of a participant, such as a fingerprint), they are not anonymous and GDPR must be applied. In brief, this stricter definition of anonymization is based on the idea that individualization can lead to reidentification. A number of studies have demonstrated how reidentification can be performed based on pseudonymized data (i.e., data on which primary identifiable information has been removed, but that do not match the criteria for anonymity of the GDPR). For instance, prior research has demonstrated successful reidentification based on magnetic resonance imaging (MRI) images using publicly available face recognition software35 but also based on face-blurred MRI images using generative adversarial networks.36 More recently, Schwarz and colleagues37 compared three types of commonly applied defacing software. Before defacing, 97% of MRI images were correctly matched to photographs using automated face recognition, while after de-facing with three popular software tools, automated face matching dropped to 28-38% of MRI scans. Beyond reidentification based on facial features, interindividual differences in brain connectomes can be matched among large datasets of brain images to effectively single out individuals,38 a phenomenon known as functional MRI fingerprinting. Ravindra and Grama39 demonstrated a de-anonymization method that utilized the uniqueness of functional MRI fingerprints to reveal the identity of individuals. We note that recent research has been more critical about the real-world risk of reidentification40 and, in practice, there is currently no consensus on the exact definition of de-identification,41 calling for additional research to better understand the privacy risks for participants.
Goal of this study
While data sharing in neuroimaging presents significant advantages, it is fundamental to take into account privacy and ethical considerations to ensure the responsible use of sensitive information. Implementing robust security measures and adhering to data protection regulations, such as GDPR, is not only a legal requirement but also essential to maintaining the trust of participants and collaborators. In practice, to share human brain images, researchers must comply with the regulations in place in the country they are based in. In the EU, this entails complying with the GDPR and its implementations across different countries.
The strict data protection and privacy requirements for anonymization in Europe pose particular challenges for data sharing. So far, there is no up-to-date inventory available that provides an overview of platforms available to EU-based researchers. In this paper, we explore the availability, awareness, and utilization of data-sharing platforms among neuroimaging researchers. We also examine the issues and challenges researchers identify with data sharing. We focus only on sharing non-anonymous (pseudonymized) brain images (sometimes referred to as “raw data”) from human participants.
Several efforts have focused on cataloging trustworthy data repositories for data sharing in research. For instance, the French Committee for Open Science (CoSO) has produced a detailed guide for selecting thematic data repositories.42 This guide helps researchers identify trustworthy repositories by using a set of seven criteria, including data moderation, the provision of persistent identifiers, and compliance with GDPR regulations. The CoSO’s evolving list[7] currently features about 50 trusted repositories, aiming to streamline the process of sharing scientific data for French researchers. Amongst those, only nine neuroimaging platforms have passed their eligibility criteria, which include deposit moderation, issuing persistent identifiers, ensuring infrastructure longevity, minimal costs, data storage inside the EU for certain data types as well as the ability to deposit regardless of institutional affiliation. Of those nine repositories, only two have been reported by CoSO as GDPR compliant. Indeed, while GDPR-compliance is a key concern for human neuroimaging, not all neuroscience data require such stringent measures. In disciplines like preclinical electrophysiology or behavioral neuroscience for instance, data sharing can be possible without maximal GDPR constraints. This, for instance, applies to a number of repositories cited for FAIR neuroscience in a recent review by Martone.43 Most of the platforms highlighted in this review for international neuroscience data sharing have data storage servers in the US exclusively. While US-based infrastructure has been instrumental in spearheading efforts towards the democratization of data sharing, it is not uniformly suitable for the sharing of human brain data by researchers from other countries (including in Europe) due to differences in legal frameworks and data protection regulations.
While data sharing is highly valuable for science, practitioners face several issues when trying to share data in support of research. In this study, we address a critical challenge in human neuroimaging research: selecting an appropriate infrastructure for data deposition. Specifically, we examine repositories suitable for sharing human brain imaging datasets by researchers based in the EU. Within the field of neuroscience, we put our focus on datasets from human participants as they require additional measures to comply with regulations making them effectively more difficult to share.
Methods
Survey
A survey (see Appendix 3) was iteratively designed and refined with input from participants of the Glioma MR Imaging 2.0 (GliMR) collaborative network, a pan-European network of experts in glioma research (see Acknowledgements for more details). This four-section survey was shared with the scientific imaging community through various platforms, including social media (Twitter[8] [now X], Mastodon, BlueSky[9]) and the GliMR newsletter and was open for contributions in the second and third quarter of 2023.
The first section of the survey queried all respondents about their familiarity with any infrastructure suitable for EU researchers to share neuroimaging data. The second section targeted respondents who answered negatively, probing whether they encountered obstacles in sharing data or faced any impediments. On the other hand, respondents who indicated awareness of relevant infrastructure were asked to provide details, including the platform’s name, URL, and whether they had previously used it to share neuroimaging data. The third section focused on participants who had already engaged in data sharing, delving into the specifics of the data types shared and whether they were linked to published papers. The final section requested participant information if they were willing to disclose such details. The Python code used to process the data from the survey is available on Gitlab[10] and archived on software heritage (swh:1:dir:a1d42e20a48847c9a7767cd7bbdd81042a71f994;origin=https://gitlab.inria.fr/empenn-public/glimr-wg2-public).
Literature review
A systematic review (drawing inspiration from44) was carried out on Google Scholar[11] to identify infrastructures referenced in the literature as suitable for storing and sharing neuroimaging data. Journal articles published before February 2025 and whose title contained the expressions ‘sharing’ or ‘FAIR’, ‘data’, and ‘neuroimaging’ or ‘neuroscience’ or ‘neuroinformatics’ were included. The words ‘platform’ or ‘service’ or ‘solution’ or ‘infrastructure’ were also required to be part of the entry’s title. Citations of articles were excluded. The exact search query run on Google Scholar is available in Appendix 4. The search results were subjected to predefined search exclusion criteria (‘abstract in a journal or conference without peer review’, ‘retracted research article’, ‘research article not dedicated to a data storage infrastructure’). The third criterion comprises research articles mentioning data sharing infrastructures but whose main claim is answering a research question or providing theoretical guidelines. Application of the search exclusion criteria was performed by visually inspecting the title and the abstract of the entries when available. Of the articles about infrastructure obtained, duplicates were removed, keeping only the last entry published, as this is more likely to represent the latest version of the platform. One review43 and the latest infrastructures list by the CoSO[12] (with domain being “biology” and subdomain “microscopy”, “neurobiology” or “bio imagery”) were added to the unique filtered research articles obtained through the search. These two resources were identified prior to the systematic search and did not appear in its results. The unique infrastructures listed in all the selected articles were extracted by carefully reading each of the articles.
Exclusion criteria
The goal of this study was to identify infrastructures that are suitable for EU-based researchers to share raw brain datasets of living human participants. We focused on infrastructures that were suitable (and had been used at least once) to share data with the community at large (i.e., sometimes referred to as “open data”). It is important to distinguish data sharing infrastructures from software. On the one hand, an infrastructure typically includes a self-hosting or cloud-based storage solution and the administrative services required to run and maintain the infrastructure. On the other hand, a software would be the tools installed on the infrastructure specifically designed and built for the purpose of sharing data. As an example, XNAT45[13] is an open source data-sharing software, and the XNAT Central46[14] was an infrastructure (decommissioned in May 2024) using the XNAT software — where users could deposit and access data. Other instances of the XNAT software may be installed elsewhere (e.g., as a lab repository).
We therefore excluded: 1) software (in the absence of an associated infrastructure) as well as 2) inactive infrastructures (i.e., discontinued or in construction as of February 2025), 3) infrastructures dedicated to other types of data (including non-brain data, non-human data, derived data or data from a predefined research project), 4) infrastructures with servers located outside of the EU and 5) infrastructures whose policy terms prevented or advised not to upload personal data or metadata.
As a note, we chose to retain infrastructures in which only members of specific institutions were allowed to upload data (i.e., download open to all but upload may be restricted; see Discussion for more on this topic).
To build a comprehensive list of data sharing platforms available to EU researchers, we assessed for eligibility all unique entries shared in the survey or identified in the literature search.
Results
Participant demographics and infrastructure awareness
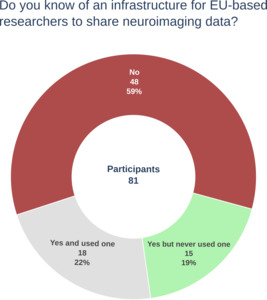
Eighty-one participants completed the survey. Of these, 40 participants (49.3%) chose to remain fully anonymous and did not provide information on their EU residency status or research involvement. Among the 41 participants (51.7%) who provided some information about their profile, all but one were based in the EU and were involved in research.
Figure 1 summarizes our main findings in terms of knowledge of an infrastructure for sharing neuroimaging data in the EU. Thirty-three participants (41%) were familiar with such infrastructure. Among them, 18 reported having previously used an infrastructure for sharing neuroimaging data covering a wide range of data types, including MRI (and derivatives, e.g., segmentations, transformation matrices), computed tomography (CT), positron emission tomography (PET), magnetoencephalography (MEG), electroencephalography (EEG), clinical and cognitive scores, and patient characteristics.
Infrastructures suitable for sharing neuroimaging data of living humans
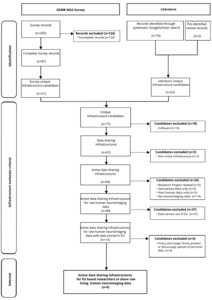
Twenty-one unique data sharing infrastructures were obtained from the survey. Nineteen research articles were obtained through a systematic literature search that described 63 unique data sharing infrastructures. By combining the infrastructures identified in the survey and those listed in the literature, we obtained 78 unique infrastructures. This initial list of infrastructures was filtered using the exclusion criteria as shown in Figure 2. 37 candidates were excluded because they did not fall within the scope of this study (10 software, 3 non-active infrastructures, 24 non-adapted infrastructures [e.g., 3 infrastructures dedicated to non-human data] and 2 only allowing the uploading of derived data). Almost three-quarters (27 out of 40) of the candidate infrastructures were then excluded because they stored their data on servers located outside of the EU. Of the 14 remaining infrastructures, 4 prohibited or advised against the uploading of personal data in their terms and conditions (see Supplementary Table 1 for the list of excluded platforms and exclusion criterion). Ultimately, 9 relevant infrastructures for the sharing of neuroimaging data from living human participants by EU researchers were obtained (see Table 1). Of those, 8 were obtained from the survey, one from the literature review and one from both the survey and the literature. This list of infrastructures is also available on Github47 (and archived on Zenodo48).
Commitments to data preservation range widely. For example, the German Neuroinformatics Node and Ebrains Virtual Research Environment ensure data preservation for ten years, while the storage guarantee for Public nEUro is contingent upon the fees paid. However, many platforms lack clear information regarding long-term preservation strategies, which could pose risks for researchers relying on these repositories. Some platforms, such as Public nEUro[15], allow datasets to be embargoed (i.e., access is restricted to reviewers until the corresponding paper is published) for a period. Six infrastructures provide Persistent Identifiers (DOI), ensuring long-term citation and accessibility of the datasets.
Challenges in data sharing for researchers
In total, 25 participants (30% of the participants) shared challenges encountered when sharing data. The respondents expressed a range of issues related to sharing neuroimaging data (see Table 2). These challenges can be broadly categorized into four key areas: legal and privacy concerns, resource and infrastructure challenges, institutional barriers, and a knowledge and awareness gap.
Legal and privacy concerns were prominent, with participants noting varying interpretations of data anonymization and identification across institutions, as well as a general lack of clarity regarding the legal landscape and relevant legislation. This uncertainty complicates the data-sharing process and may deter researchers from engaging in data sharing altogether.
Resource and infrastructure challenges were also highlighted, particularly issues related to handling large datasets, the complexities of existing payment models, and the high costs sometimes associated with long-term data archiving. For example, sharing data using Dataverse NL requires institutions to pay a membership fee per year of €5,950 plus value added taxes (VAT) in 2025, and an additional cost of €330 plus VAT per terabyte (TB) per year (see https://dans.knaw.nl/en/data-services/dataversenl/). Despite efforts to address these challenges,49 many participants expressed frustration with the lack of adequate time, incentives, and resources to develop robust data-sharing systems that are safe and sustainable over the long term.
Institutional barriers further impede data sharing. While respecting data privacy and ethics is essential to the research process, in practice they sometimes translate into lengthy administrative processes.50 Institutional inertia, such as slow data use agreement approvals along with reluctance with those in leadership, including Principal Investigators, were also cited as significant obstacles.
Knowledge and awareness gap among researchers. Many participants were unaware of available data repository solutions and the possibilities for establishing secure data access procedures. There was also variability in how different institutions interpret the same data in terms of anonymization, pseudo-anonymization, or identifiability, which adds another layer of complexity to the data-sharing process.
Discussion
The results of this survey provide insights into the current landscape of neuroimaging data sharing in the EU. It is notoriously difficult for EU-based researchers to identify an infrastructure that is suitable to share human neuroimaging data. This is well demonstrated by the fact that less than half of the respondents were aware of a data-sharing infrastructure and only about 20% had used one previously. Of note, the literature review only made it possible to identify two infrastructures. In total, we identified nine infrastructures suitable for sharing brain imaging data from human participants for stakeholders based in the EU. This list of infrastructures is also available on Github47 (and archived on Zenodo48) to support further updates from the community. Many of the available repositories have the geographic locations of their servers in the US, raising potential issues for compliance with European data protection laws. While platforms like the German Neuroinformatics provide guarantees regarding sustainability, others lack clear information on long-term data preservation commitments. Some repositories, like Sikt, impose relatively high fees on data uploads, which may limit access for some researchers, while others, like the EBRAINS Virtual Research Environment, offer long-term storage by relying on institutional support.
As a note (and in departure from other reviews such as51), we chose to retain infrastructures in which only members of specific institutions could upload data (i.e., download open to all but upload may be restricted). We believe that it is important to also highlight such platforms for two main reasons: 1) Although they cannot be used by all to upload data (as data upload is restricted to members-only), these infrastructures are open-to-all for data download. This effectively makes them relevant for the community at large and, in particular, for those interested in data reuse; 2) More importantly, this model – i.e., data sharing infrastructures in which members-only can upload but all can download – seems to be widespread in the EU. As an illustration, out of the nine platforms identified, only four offered upload open for all. We believe this is likely to expand as more institutions are encouraged to provide such platforms for their own scientists.
Despite these advances, perceived and administrative barriers to data sharing remain high. The complexities of compliance, coupled with logistical and institutional hurdles, continue to impede the seamless exchange of data. Our survey revealed several challenges that researchers encounter when trying to share data, highlighting four key areas for improvement. First, it is important to clarify the legal and privacy frameworks. Establishing standardized definitions of data anonymization and identification across institutions can reduce confusion regarding compliance with relevant legislation. Second, investing in infrastructure and resources is crucial. This includes developing cost-effective solutions for handling large datasets and simplifying payment models for data storage and archiving. By providing adequate time, incentives, and resources for building sustainable data-sharing systems, institutions can foster a more collaborative environment. Third, it is essential to address institutional barriers. This may include streamlining the ethics approval process but also providing researchers with appropriate data use agreements (see the Open Brain Consent52 for a set of templates that may be used as a basis). Institutions should also advocate for clearer guidelines on data sharing that empower researchers while ensuring regulatory compliance. Funding agencies should continue to support and develop stable databases to help take storage and sharing burdens off the shoulders of individual research institutions.53 Finally, bridging the knowledge and awareness gap is necessary. Increasing education and training on available data repository solutions and secure data access procedures will enable researchers and data managers to utilize these tools effectively. By fostering a better understanding of data-sharing practices and addressing the variability in interpretations of data identifiability, researchers will be better equipped to engage in collaborative efforts. Additionally, addressing these challenges will only be possible by creating incentives for data sharing. As an example, the development of data papers – a format allowing researchers to describe and publish their datasets as a citable publication – (see for example54) is a step in this direction. Beyond that, further efforts are needed to make data sharing valued as a component of scientific contributions. Finally, providing funding and support to researchers to cover the needs associated with preparing data for sharing is a requirement to encourage more widespread participation. These results are in line with the earlier survey conducted by the European Cluster for Imaging Biomarkers17 in particular, on highlighting concerns about the time and human resources required for data sharing but also regarding the administrative and legal barriers. In contrast, fears of being pre-empted (or “scooped”) was not mentioned as an issue related to data sharing in the current work. Overall, the survey findings underscore the need for clearer guidelines, improved infrastructure, and enhanced education to support more effective and widespread sharing of neuroimaging data among researchers.
It is important to recognize that our work primarily focused on the sharing of image data, which may be a limitation. In practice, image data must be accompanied by various types of metadata to be truly useful and are often complemented by other data such as questionnaires, physiological samples, psychological or diagnostic tests. In practice, the addition of further data and metadata often complexifies the assessment over the risk of reidentification.55 In addition, we note that our estimated percentages of researchers knowing about or having used a data-sharing infrastructure is likely an overestimation. Indeed, the survey was shared using neuroimaging open science channels effectively biasing the sample towards professionals already engaged in or interested in open science. As a proxy for GDPR-compliance, we excluded infrastructures whose policy terms were explicit in preventing or advising users not to upload personal data or metadata as well as infrastructures with servers located outside of the EU. While those two features are essential for GDPR-compliant data sharing, we note that data protection officers should always be consulted before choosing and uploading data to a platform.
While data sharing is an important lever to build more heterogeneous samples and therefore limit bias in downstream models, we note that the fairness of predictive models is a research topic in itself that goes well beyond the question of the diversity of the input datasets.56 In addition, sampling inequalities in neuroimaging studies57 has a strong impact on the studied population that data sharing alone will not be able to counter.
Despite the promising frameworks and support for data sharing, practical implementation is fraught with challenges. The scientific community faces conflicting expectations: while open science policies and international scientific cooperation demand extensive data sharing and pooling of smaller studies for meta- and mega-analyses to achieve broader perspectives and more precise conclusions, the GDPR emphasizes personal data minimization and privacy protection. This discrepancy sometimes creates confusion amongst researchers which can lead to reluctance in sharing data. Inter and intra-country variations in data management practices further complicate the situation. Focusing on these issues in future research could pave the way for establishing a European consensus on best practices for neuroimaging data sharing. This could help mitigate inconsistencies and promote uniform standards across institutions, ultimately facilitating more effective and compliant data sharing practices.
Conclusion
This work has outlined the unique challenges and complexities associated with sharing brain imaging data in a GDPR-compliant environment. In total, we identified nine data sharing infrastructures, suitable for researchers based in the EU to share brain imaging datasets. The survey highlighted the lack of awareness among researchers about the available platforms for sharing imaging data, underlining the urgent need for clearer guidelines and enhanced educational efforts. In addition, we have identified legal and privacy concerns, resource and infrastructure challenges, institutional barriers, and knowledge and awareness gaps as four key obstacles to effective data sharing.
The future of neuroimaging data sharing hinges on publicizing existing GDPR-compliant infrastructures as well as fostering new robust and GDPR-compliant solutions that facilitate both data protection and accessibility. Future efforts should focus on building cross-border consensus on best practices, enhancing awareness, and developing incentives for data sharing. Funding bodies and research institutions must play a crucial role in supporting initiatives that provide stable, long-term repositories aligned with FAIR and CARE principles.
Author Contributions
Lefort-Besnard Jérémy: Conceptualization, Writing-original draft, Data curation, Formal analysis, Visualization, Validation, Writing-review & editing. Pron Alexandre: Data curation, Methodology, Formal analysis, Visualization, Validation, Writing-original draft, Writing-review & editing. Clement Patricia: Conceptualization, Resources, Writing-review & editing. Barker Gareth: Writing-review & editing. Erez Yaara: Formal analysis, Supervision, Writing-original draft, Writing-review & editing. Fernández-Seara Maria A.: Conceptualization, Writing-original draft, Writing-review & editing. Feyzioglu Saide Begüm: Writing-review & editing. Hartman Shir: Data curation, Formal analysis, Writing-original draft. Iglesias-Rey Ramón: Conceptualization, Investigation, Writing-original draft, Writing-review & editing. Keil Vera: Resources, Data curation, Validation, Investigation, Visualization, Writing-review & editing. Mitrović Tatjana: Writing-original draft, Writing-review & editing. Nechifor Ruben Emanuel: Conceptualization, Visualisation, Writing-review & editing. Ozturk-Isik Esin: Conceptualization, Writing-original draft, Writing-review & editing. Passarinho Catarina: Investigation, Writing-original draft, Writing-review & editing. Petrušić Igor: Investigation, Writing-original draft, Writing-review & editing. Šerifović Trbalić Amira: Writing-original draft, Writing-review & editing. Sollmann Nico: Conceptualization, Data curation, Formal analysis, Validation, Investigation, Methodology, Writing-original draft, Writing-review & editing. Thust Stefanie C.: Conceptualization, Writing-original draft, Writing-review & editing. Turhan Gülce: Writing-original draft, Writing-review & editing. Warnert Esther: Conceptualization, Data curation, Funding acquisition, Investigation Writing-original draft, Writing-review & editing. Heunis Stephan: Writing-review & editing. Maumet Camille: Conceptualization, Methodology, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Data and Code Availability
The Python code used to process the data from the survey is available on Gitlab[16] and archived on software heritage (swh:1:dir:a1d42e20a48847c9a7767cd7bbdd81042a71f994;origin=https://gitlab.inria.fr/empenn-public/glimr-wg2-public).
The raw and processed survey data are not publicly available due to the presence of personally identifiable information, for which explicit consent for public sharing was not obtained.
Funding Sources
Jérémy Lefort-Besnard was supported by Région Bretagne (Boost MIND) and by Inria (Exploratory action GRASP). Alexandre Pron was supported by the IAM hub of France Life Imaging (grant ANR-11-INBS-0006). The Applebaum Foundation provided partial financial support in the form of a grant award to Yaara Erez. The Ministry of Innovation, Science and Technology, Israel, provided partial financial support in the form of a grant award to Yaara Erez (No. 0005791). Ramón Iglesias-Rey (CP22/00061) from the Miguel Servet Program of Instituto de Salud Carlos III and Co-financed by the EU. Esin Ozturk-Isik and Gülce Turhan were partly supported by TUBITAK 115S219 and 216S432 grants. Catarina Passarinho was supported by FCT - Fundação para a Ciência e Tecnologia, I.P. by grant 2022.13185.BD (https://doi.org/10.54499/2022.13185.BD) and LARSyS-FCT grants 10.54499/LA/P/0083/2020, 10.54499/UIDP/50009/2020, and 10.54499/UIDB/50009/2020. Igor Petrusic is supported by the Ministry of Science, Technological Development and Innovation, Republic of Serbia (contract number for IP: 451-03-136/2025-03/200146). Stefanie Thust receives research funding support from the NIHR and Cancer Research UK. As an employee at Research Centre Jülich, Stephan Henuis at the institute for neuroscience and medicine (INM-7), developed software with support from: the German Federal Ministry of Education and Research (BMBF 01GQ1905), the US National Science Foundation (NSF 1912266), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under grant SFB 1451 (431549029, INF project), the MKW-NRW: Ministerium für Kultur und Wissenschaft des Landes Nordrhein-Westfalen under the Kooperationsplattformen 2022 program, grant number: KP22-106A.
Acknowledgements
This work was undertaken as part of Glioma MR Imaging 2.0 (GliMR) supported by European Cooperation in Science and Technology (COST) Action CA18206[17] (2019-2024) and by ESMRMB as a Working group (2024-ongoing). GliMR aims at creating a pan-European network of experts in glioma research, including patient organizations, data scientists, and MRI specialists.13 The work carried out in this manuscript was more specifically led by COST Action CA18206 Working Group 2 (WG2) “multi-site data integration”. The authors extend their thanks to the respondents of the survey as well as to those who facilitated its dissemination.
Conflicts of Interest
Alexandre Pron was a contributor to the Shanoir software (see Shanoir contributor list) as part of his work funded by FLI-IAM (see Fundings section). Camille Maumet was part of the leadership of the Shanoir software as president of the Executive Committee of the Shanoir Consortium. The Shanoir Neurinfo infrastructure managed by the Neurinfo platform is regularly used in research projects of the Empenn team of which Jeremy Lefort-Besnard, Alexandre Pron and Camille Maumet were a part. Vera Keil joined the EUCAIM consortium in March 2025 with her institution. EUCAIM glioma data is shared via XNAT/Health-RI, but Vera Keil does not have an official function in XNAT. Stephan Heunis developed the open source software tool datalad-catalog that the Public nEURo system uses as the frontend for findability and accessibility of the datasets that they host. However, he is not involved in the Public nEURo project in any other way shape or form, they just use the software that he created at his current employment with funding support from several sources (see Funding Sources section).