Introduction
Open, large-scale, inclusive datasets - everyone wants them! Such datasets are especially critical in clinical neuroscience research to substantiate clinically relevant markers and models of disease. Yet finding and reusing clinical datasets comes with unique challenges. Over the last two decades, significant efforts in promoting open science practices and data sharing have been instrumental in advancing the field of neuroimaging.1–4 These practices have been codified in the FAIR principles5 that advocate for outputs of scientific work to be Findable, Accessible, Interoperable, and Reusable to ensure impact and reuse of data, and foster efficient scientific collaboration. Open data infrastructure,3,6 community standards,7,8 and tools9–12 have emerged that implement these principles and support collaboration and sharing of research efforts at scale.
But not all data can be openly shared. For large scale collaborations such as the ENIGMA consortium,13,14 the adoption of open science principles is often complicated by a variety of local constraints relating to tools, training, research culture, privacy, legality, informed consent and data governance. Differences in constraints across sites, particularly in local data governance and privacy requirements such as the European General Data Protection Regulation15 or the Canadian Personal Information Protection and Electronic Documents Act,16 make the adoption of existing open standards, processes, and centralized platforms extremely difficult. Instead, sites resort to ad-hoc, siloed solutions that often lead to divergent data organization and processing practices and create coordination challenges.
The different needs of research consortia therefore require a targeted approach to embed FAIR principles – built around decentralized neuroinformatics infrastructure and tools, which promote open and reproducible science while respecting local data governance and privacy requirements. Additionally, such an approach has to emphasize the ease-of-use and intuitiveness of these tools to account for the heterogeneous technical experience of users in a clinical context. We believe that the adoption of such a decentralized open science infrastructure is particularly crucial in an era where a growing majority of datasets require access control.
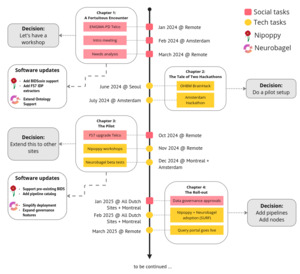
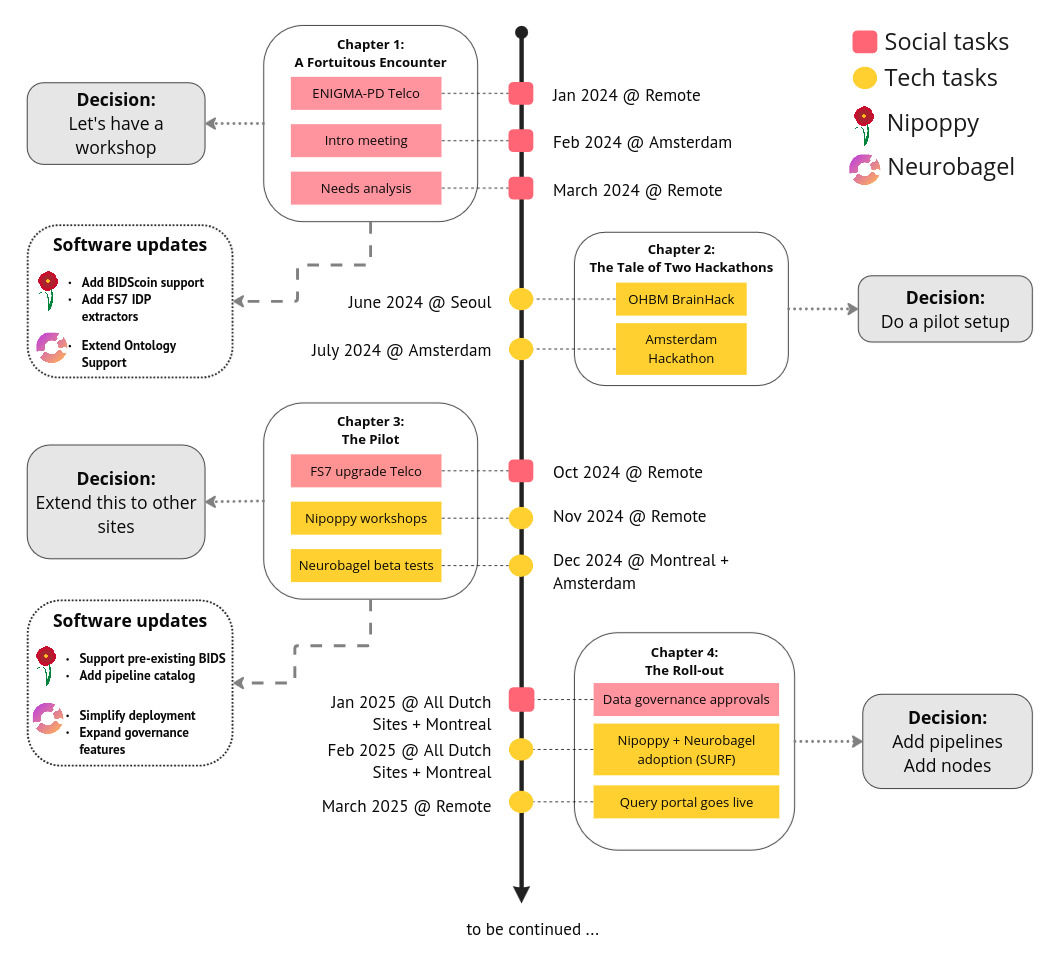
Here we report on the outcomes of a recent pilot project (see Figure 1) we conducted to deploy a decentralized neuroinformatics architecture in a large research consortium to facilitate the adoption of open science principles, in particular the findability of access-controlled data. We conducted this pilot project as a collaboration between the ENIGMA Parkinson’s Disease (ENIGMA-PD) working group (PD-WG, https://enigma.ini.usc.edu/ongoing/enigma-parkinsons/), and the neuroinformatics team from the ORIGAMI lab at McGill University in Montreal (https://neurodatascience.github.io/). ENIGMA-PD is a large research consortium comprising more than 40 international sites that is coordinated from the Amsterdam University Medical Center (UMC) and the University of Southern California (USC). Because of the internationally distributed nature of the PD-WG, the ability to search and find data across participating sites is an important, and initially unmet, need to assess the feasibility of future research projects. We describe the challenges we encountered in addressing this need, our solutions, and the achieved outcomes, in the hope that other groups can build on our experience in navigating similar challenges in open science adoption. We group the challenges into two categories. The technical challenges: 1) data standardization, 2) consistent processing, 3) semantic harmonization, and 4) managing of cloud services; and the social challenges: 1) ease-of-use, 2) training, and 3) data governance. Addressing these challenges required innovation, patience, and at times incentivization of users through gamification. Through these efforts, we have successfully created a decentralized data platform that enables subject-level discovery of more than 1200 subjects with multimodal data across access-controlled datasets from ENIGMA-PD sites in Europe and Canada.
Teams and Tools
The ENIGMA Parkinson’s Disease working group (PD-WG) is the largest international neuroimaging effort to advance our understanding of this progressive neurodegenerative disease by combining data from 44 participating research centers across five continents. The PD-WG is coordinated by a team of researchers from Amsterdam UMC and USC as part of the ENIGMA consortium13 which embraces a collaborative and inclusive approach that brings together researchers from around the world, including those from lower-resourced countries. The PD-WG regularly holds online meetings to discuss projects and coordinate protocols. Participation in projects involves sharing of processed data (i.e., derived data from individual participants) with the coordinating site, to facilitate centralized mega- and meta-analysis. Researchers with relevant data, methods or project ideas are welcome to join and to securely access approved data. This collaborative model has been very productive and has led to several high-profile publications by the PD-WG (see https://enigma.ini.usc.edu/ongoing/enigma-parkinsons/).15–19
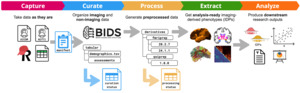
The ORIGAMI lab at McGill University in Montreal has long standing expertise in developing neuroinformatic tools and platforms and has contributed to many open science initiatives and their maintenance over the years. Inspired by the ReproNim effort20,21 and building on ReproNim tools,22,23 the lab developed two complementary tools to improve research FAIRness: Nipoppy24 and Neurobagel.25 Nipoppy is a lightweight framework for standardized organization and processing of neuroimaging datasets. Neurobagel is an ecosystem for distributed dataset harmonization and discovery. Together, these tools can facilitate coordination of data curation, processing, and discovery across multiple sites.
Nipoppy (see Figure 2) serves as a “dry-lab protocol” accompanied by a Python package that guides researchers from raw data to analysis-ready imaging-derived phenotypes in a reproducible manner. By simplifying the use of specific pipeline versions and their run-time parameters, it helps ensure consistency, tracks outputs, and automates updates when new subjects are added or data are reprocessed. It is built around existing community standards for data organization, such as the Brain Imaging Data Structure (BIDS),7 and technologies for reproducible processing such as containers and Boutiques.26 The accompanying Nipoppy Python package (https://pypi.org/project/nipoppy/) provides a user-friendly interface for setup, upgrades, and tracking of neuroimaging pipelines.
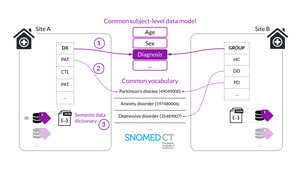
Neurobagel is an ecosystem of tools that enables subject-level search across decentralized datasets without the need for access to the underlying data. Tabular, non-imaging data like demographics and clinical assessments are often named and structured in idiosyncratic ways, making cross-dataset cohort discovery very challenging. The graphical Neurobagel annotation tool allows researchers to map these local variables to standardized FAIR vocabularies (e.g. “Age”, “Sex”, “Diagnosis”, “Assessment”) and stores them as data dictionaries. The command line interface (CLI) then uses these mappings to generate standardized availability records which can be combined with imaging and derivative modalities about the same dataset. The annotation of local data allows for standardized data availability records to be automatically extracted without having to rename or change the raw data.
Once standardized availability records are generated, each site locally deploys a Neurobagel discovery node that consists of a lightweight web application programming interface (API), and an internal graph database. The API can respond to queries with varying levels of detail depending on the local governance, from a binary response (“we have data like that”), to aggregate counts (“56 subjects match your query”), or individual harmonized data points. This allows sites in the PD-WG to make local data findable while complying with their local data governance and ethics requirements.
Chapter 1: A fortuitous encounter
The teams from ENIGMA-PD and the ORIGAMI lab met each other for the first time at the monthly ENIGMA-PD virtual meeting in January 2024. One of the topics on the agenda was the upgrade of the FreeSurfer pipeline27 from version 5 to 7 and the subsequent extraction of segmentations. This upgrade had been planned for about two years as part of the new image processing protocol for ENIGMA-PD sites, but ensuring a consistent rollout across all sites had proven challenging. The sites have very limited time and resources to commit to data processing requests from the PD-WG. During this call, the ORIGAMI members realized their tools would be a great fit for addressing these challenges, reached out to the ENIGMA-PD team offline, and made plans to meet in Amsterdam (February 2024) to better understand the needs of the PD-WG (see Figure 1).
The initial meeting in Amsterdam highlighted the considerable effort required to manage a large, global consortium and the technical and social challenges associated with it. Without dedicated data infrastructure, ENIGMA-PD workflows diverged across sites and projects, creating inefficiencies at every step – from adding a new site, establishing data availability, managing processing tasks, to pooling processed data. For instance, establishing phenotype availability currently requires repeated surveys and manual searches across sites. Limited local resources further complicate coordination.
Within the broader goal of simplifying end-to-end data management, we prioritized two immediate problems: 1) expediting the PD-WG-wide reprocessing with an updated FreeSurfer version and 2) facilitating discovery of available data (raw and processed) to design and assess the feasibility of new projects. These were central use cases for which Nipoppy and Neurobagel were developed.
Based on these initial discussions, the Amsterdam team tested the tools independently (April-May 2024), while the ORIGAMI team improved documentation and added support for additional BIDS converters (e.g., BIDScoin). Plans were also made to organize two workshops for training members from multiple sites.
Chapter 2: The tale of two hackathons
Between June and July 2024, we organized two hackathons to understand existing data workflows within ENIGMA-PD and their current limitations. Our goal was to propose the adoption of Nipoppy and Neurobagel as a long-term investment in neuroinformatics infrastructure that would not only help with the planned FreeSurfer 7 upgrade but also offer improvements in the efficiency and FAIRness of data processing and analysis for the PD-WG sites.
Hackathon 1: BrainHack, Organization for Human Brain Mapping (OHBM), Seoul (Dates: June 2024).
Attendees: Sites from Europe and East Asia.
Challenges: For the ORIGAMI group this was the first collaboration with a large, clinical consortium. The heterogeneity of imaging protocols and naming practices meant that sites often required one-on-one support, and so a larger than expected portion of the hackathon was spent on data standardization. Different levels of prior technical experience among attendees meant that extensive training on prerequisites such as the use of virtual environments, containers, Git, etc. was needed before moving on to open science tools and standards. In this phase we encountered questions on what BIDS is and how to perform BIDS conversion for less common formats (PAR/REC data, multiple field maps, 7 Tesla, multi-echo imaging, etc).
Outcomes: The majority of time at the hackathon was spent on helping sites organize their existing data according to the BIDS standard using Nipoppy. By the end, three out of five participating sites had successfully initialized Nipoppy and were able to launch a test pipeline.
Lessons: The heterogeneity of imaging protocols and naming practices makes BIDSification a labor-intensive step that needs expert support. In person workshops are very helpful to help overcome this initial barrier before users become stuck or discouraged.
Hackathon 2: Amsterdam UMC, Amsterdam, The Netherlands (Dates: July 2024).
Attendees: ENIGMA-PD core group, and trainees and early career researchers (ECRs) involved in the ENIGMA-PD-WG from Amsterdam, Nijmegen and Groningen, The Netherlands.
Challenges: The Dutch sites possessed more experience and expertise in data curation and processing tasks, so the primary challenges we encountered were related to the scale of their datasets and to figuring out a transition plan from the existing local setup to one involving Nipoppy and Neurobagel in order to establish consistent FreeSurfer processing, tracking and discovery of its outputs across sites.
Outcomes: Two out of the three Dutch sites were able to adopt the Nipoppy recommended workflows and initiate processing of their data with FreeSurfer 7. All sites were also able to try out the Neurobagel tools in a sandbox (i.e. local machine) which allowed us to demonstrate cross-site discovery of harmonized demographic and imaging data through Neurobagel to the hackathon participants, in particular to the principal investigators of each site.
Lessons: In-person workshops are highly effective for both technical and social challenges. Practical demonstrations, such as cross-site discovery using hackathon data, help users appreciate the value of discovering data before access. The privacy constraints on various data types (e.g. demographics, raw versus processed imaging) were unclear, requiring further institutional discussions before sites could opt-in into public dataset discovery.
The two hackathons provided important insights into the utility and feasibility of the proposed infrastructure adaptation of Nipoppy and Neurobagel across ENIGMA-PD sites. For the next steps, we opted for a two-pronged approach involving: 1) a comprehensive pilot project within the Dutch and Canadian sites to deploy the entire neuroinformatics stack of Nipoppy and Neurobagel tools and 2) a more focused initiative with the larger ENIGMA-PD consortium to encourage Nipoppy adoption for their FreeSurfer 7 upgrade process.
We initiated both efforts in parallel and planned to make use of the available institutional support at the Dutch sites for the more ambitious technical deployment in the pilot project, while incrementally rolling out deployment of Nipoppy to global sites with limited availability of resources. We expected that the practical experiences gained from the pilot project would then help us refine our tools and the deployment plan that depends on institutional buy-in.
Chapter 3: The pilot project and the gamification strategy
Although we ultimately wanted to enable standardized processing and data discovery across all ENIGMA-PD sites, we focused our pilot project on four sites in the Netherlands and Canada to test our assumptions before a wider roll-out. The existing collaboration between the pilot sites offered an easier access to the institutional infrastructure and decision makers for data governance, which we expected to be critical especially for the data discovery aim of our project.
Given the time sensitive nature of FreeSurfer 7 processing for the PD-WG, in parallel we launched an effort to deploy Nipoppy at all international PD-WG sites. Nipoppy can be deployed independently of Neurobagel and has fewer infrastructure and data governance requirements. Focusing on standardized processing with Nipoppy thus allowed international sites, where access to institutional infrastructure was less certain, to benefit from technical support and a user-friendly process to adopt the new processing pipelines for the PD-WG.
We aimed for achieving Nipoppy adoption at several sites (4 pilot and 10 global) and cross-site search for the pilot sites within the same timeframe (~6 months).
Standardizing data curation and processing with Nipoppy
Target audience: All sites (pilot + global).
Goal: Help sites run their processing tasks (FreeSurfer 7 upgrade and downstream segmentation) through Nipoppy setup.
The FreeSurfer upgrade plan was officially announced at the PD-WG call in October 2024. We briefly explained the motivation and benefits of Nipoppy, and sites were strongly encouraged to adopt Nipoppy for this purpose. To support this rollout, the ENIGMA core team together with Nipoppy developers organized multiple online workshops (November 2024) and set up a Discord server for asynchronous tech support. In order to track the adoption process and as a social incentive to encourage adoption, we created a simple leaderboard where the sites could update their progress.
Challenges: Common technical challenges in this phase were: 1) creation and validation of heuristics for BIDS conversion, 2) installing and working with software containers (Apptainer), 3) setup, data transfer, and working on high-performance computing (HPC) systems.
Enabling cross-site search at the pilot sites with Neurobagel
Target audience: 3 Dutch and 1 Canadian site.
Goal: Enable search of available tabular and imaging data (BIDS + FreeSurfer 7) across 4 sites.
This pilot project built on the rapidly progressing data curation and processing efforts at the PD-WG sites in Amsterdam, Nijmegen, Groningen, and Montreal to enable data discovery across these sites with Neurobagel.
Our specific aims for the pilot project were to:
-
Ensure that at least 3 geographically distinct institutes deploy one discovery node each with at least one dataset each (see Table 3).
-
Demonstrate the ability to search across these discovery nodes, including on preprocessed data availability (e.g. FreeSurfer 7), in line with each institute’s data governance requirements.
-
Assess whether sites can be trained to maintain and update their own nodes incrementally as more data becomes available.
Challenges: The initial deployment plan expected the discovery nodes to be hosted within each institute and maintained by the local IT personnel. This quickly proved infeasible because the IT policy of the Amsterdam University Medical Center (UMC) prohibits research-focused services to be deployed internally. We adjusted the plan to instead have sites deploy their Neurobagel nodes on the research cloud infrastructure from SURF, the IT cooperative of Dutch education and research institutions. All Dutch sites could obtain access to this research cloud (the site in Montreal had access to equivalent services from the Digital Research Alliance of Canada). This change of plans meant moving the data availability information (metadata) from the local institute to the SURF cloud, which required additional data governance approvals. Because the research cloud virtual machines were provided without dedicated IT support, each site had to deploy and maintain their nodes on their own. We asked each site to designate one ECR who we would train and support in this task.
At this stage, the two parallel initiatives, i.e. the consortium-wide push for the Nipoppy adoption and the more focused pilot effort to achieve data discovery with Neurobagel helped us gain practical experience with the decentralized deployment of our tools and identify technical and governance challenges early on in the process. We used this experience to formalize a self-sustaining rollout process that would ensure standardized data workflows at all sites enabling continuous data discovery and sharing, for current and prospective datasets.
Chapter 4: The rollout
The rollout spanned the period (January-April 2025) of decentralized adoption of the tools and was primarily supported via asynchronous communication platforms (Discord, Slack, Github). We saw this as a real-world validation of our overall strategy and planning aimed at organic growth and adoption of our open tools. We were supporting Nipoppy adoption at 14 sites (4 pilot and 10 global) and the additional deployment of Neurobagel to enable cross-site search for the 4 pilot sites.
Standardizing data curation and processing with Nipoppy
During the rollout period, the sites relied on Discord servers and the documentation resources from the core Amsterdam team (https://github.com/ENIGMA-infra/ENIGMA-PD) and Nipoppy (https://nipoppy.readthedocs.io/en/stable/index.html) for setting up Nipoppy tools to help with FreeSurfer upgrade and related tasks. Thanks to earlier hackathons and workshops, we now had a growing group of ECRs from the Montreal and Amsterdam teams with neuroinformatic expertise who were able to provide support to the larger ENIGMA-PD community. The leaderboard was updated by the sites and reviewed periodically at the ENIGMA-PD WG meetings, which seemed to be positively reinforcing adoption.
Challenges: Although the lessons from hackathons and the efforts put into initial planning helped considerably with the rollout, the larger scale of a full deployment raised a few new challenges. These were discussed and resolved on the Discord server. We list some of these below in Table 4.
Outcomes: Through the community effort, we resolved these challenges and made rapid progress. By mid-April 2025, 17 datasets/sites had completed BIDS conversion, and 15 had successfully run FreeSurfer 7 preprocessing using the Nipoppy framework. Due to closer collaboration, the Dutch and Montreal sites made rapid progress and quickly moved onto Neurobagel deployment for data discovery.
Enabling cross-site search at the pilot sites with Neurobagel
After testing the deployment plan at Amsterdam UMC, in January 2025, we invited the other Dutch sites to join the planned Neurobagel pilot project. All sites had attended at least one of the preparatory hackathons and were actively engaged in the parallel international rollout of Nipoppy, and were therefore highly motivated to address the data discovery challenges in the PD-WG by participating in the pilot.
We began by reaching agreement on the granularity at which each site would be able to make local data discoverable based on local data governance requirements. All sites agreed to allow an aggregated discovery of their local data that returns only the total number of participants matching a cohort, but no individual details about these participants. Additionally, the sites asked that cohort results below a certain minimum number of participants would be censored to prevent very specific queries that could identify individual participants. The ORIGAMI team developed this “minimum cell size” feature in a new release of Neurobagel and all sites selected a locally appropriate cutoff level for their node.
To better evaluate the feasibility of a future rollout of Neurobagel to all PD-WG sites, we set ourselves an ambitious timeline of completing the pilot project for all four sites in 2 months. As a shared external commitment, we proposed to tie the pilot project to the submission of a case report to this Aperture special issue, giving us a helpful and tight external deadline. Each site nominated at least one ECR to lead the local data preparation and Neurobagel node deployment, and we sought to continue the fun and collegial atmosphere of the hackathons to facilitate the progress. In this sense, our approach to the Neurobagel pilot project was a test of a possible strategy for the later, wider rollout of the data standardization and discovery efforts across the PD-WG.
Each site began the process of preparing their local data (see Figure 3) for discovery by 1) identifying a tabular file with subject-level demographic and phenotypic measures (or by combining existing files), 2) annotating the tabular file with the graphical annotator tool, and then 3) extracting availability information with the Neurobagel CLI. As an additional step, our revised deployment plan also required each site to deploy and maintain their local Neurobagel discovery node on the SURF research cloud infrastructure. As one purpose of the pilot project was to test the feasibility of an eventual expansion of Neurobagel and Nipoppy to the remaining PD-WG sites, we decided to limit our support to guidance and training in the form of online meetings and documentation, with ECRs doing the actual work themselves. We scheduled an initial online meeting with all sites to explain the process, identify challenges and offer support, and follow-up meetings with individual sites for annotation and deployment.
We encountered three main challenges in this process: 1) Most sites struggled to find good matches for their locally acquired assessments in the controlled vocabulary Neurobagel supported for this purpose. In response, the ORIGAMI team replaced the vocabulary with a more clinically focused one (SNOMED-CT) that provided better coverage of the tools used by the PD-WG sites and made the update available to all users. 2) Some sites used different participant IDs for imaging data and tabular/phenotypic data, making it impossible for the Neurobagel CLI to link information about the same participant. We used the online meetings to help sites add a new column to their tabular data and create a mapping with the existing BIDS participant IDs, allowing all sites to successfully extract local data availability records. 3) An unexpected challenge was helping ECRs connect to their SURF cloud machine via the SSH protocol to transfer data availability records and deploy their node. Most ECRs used Windows workstations that needed dedicated software and documentation for these steps.
A member of the Montreal team who was familiar with Windows wrote a step-by-step guide that also made use of the excellent documentation provided by SURF itself. The ORIGAMI team also simplified the Neurobagel node deployment process into a three-step, semi-automatic recipe that included everything from the node services themselves, automatic data loading, a self-configuring reverse proxy and automatic provisioning of SSL certificates, leaving only the configuration and launching of the node as manual steps. The simplified recipe worked well, as all sites were able to deploy their nodes during a single Zoom call.
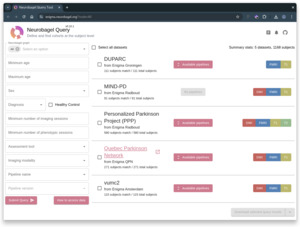
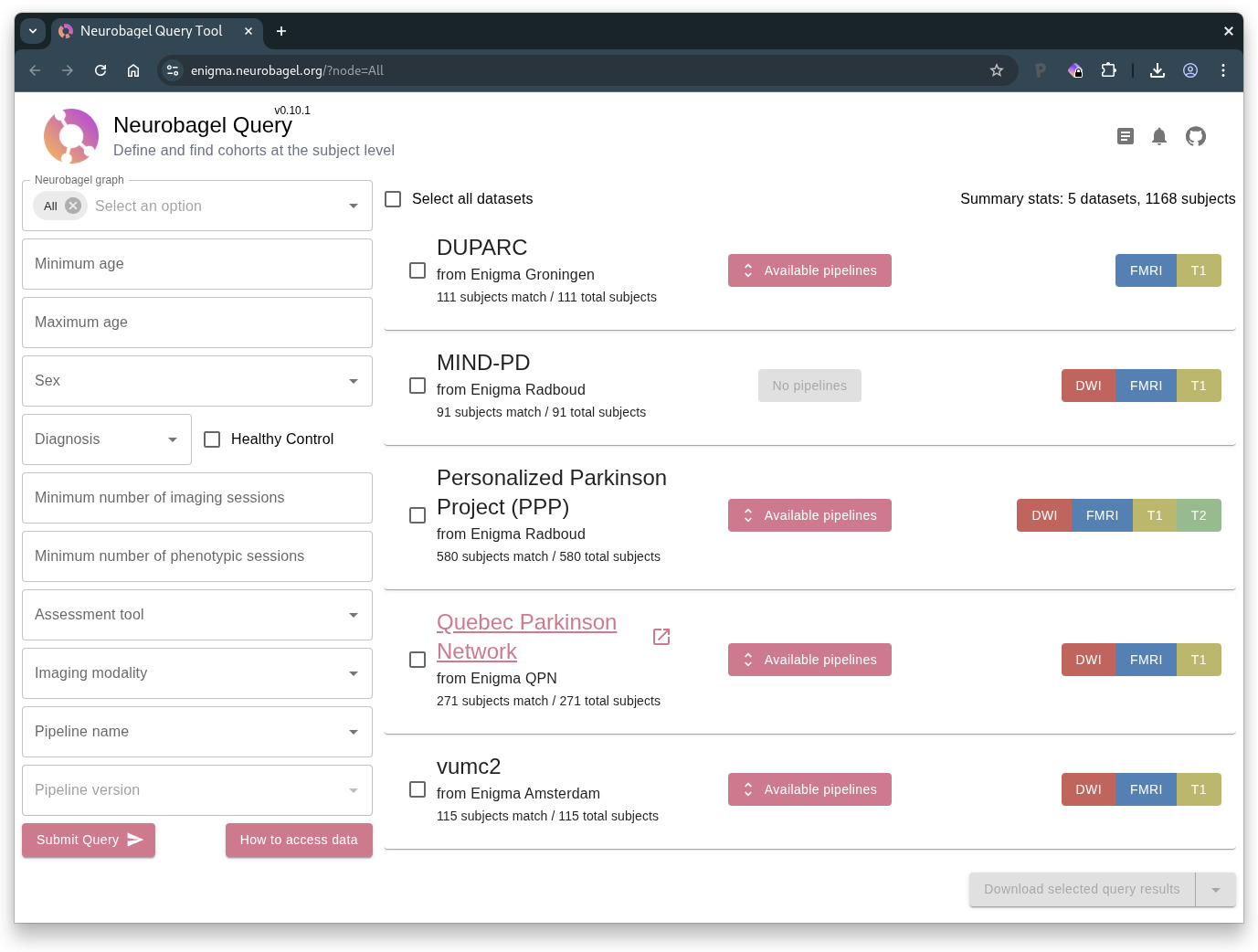
Outcomes: By March 2025, all four sites were running a Neurobagel discovery node with at least one dataset, and a total of more than 1000 participants. We deployed a federation portal where interested researchers can search for cohorts across these PD-WG datasets that match specific inclusion criteria.
Discussion
Since our initial meeting a year ago, our teams have pursued two parallel efforts to help the ENIGMA-PD working group address current challenges inefficient data processing: 1) The standardization of data curation and processing with Nipoppy across all international ENIGMA-PD sites, and, building on this effort, 2) a pilot project to achieve cross-site cohort discovery with Neurobagel across three Dutch and one Canadian site of ENIGMA-PD.
The benefits
Both efforts have made rapid progress. Between December 2024 and the time of writing in April 2025, 17 of the 42 international ENIGMA-PD sites, including all four sites involved in the discovery pilot project, have successfully deployed Nipoppy and are using it to process their data with the new Freesurfer 7 version required for future projects. In parallel, two months after beginning the cross-site discovery pilot project, all four pilot sites have successfully harmonized their local data and deployed their Neurobagel discovery nodes, allowing us to demonstrate a functional cohort discovery across local data from four ENIGMA-PD sites: Neurobagel Query Tool. At the time of writing, 5 datasets with a total of over 1,000 participants are queryable, including on the availability of imaging derivatives preprocessed with Freesurfer and other pipelines (Figure 4). The pilot project has achieved its key objectives and has helped us get a better sense of challenges and of promising strategies to address them, that will help guide our future plans to expand this technology to other sites in ENIGMA-PD.
For the ORIGAMI development team behind Nipoppy and Neurobagel, this project has been very helpful. For tool developers it can be difficult to receive honest feedback on what struggles or concerns users face when using a tool, and whether it solves an important problem for them. Through working in person with a group of motivated users, we understood where in the process they got stuck or were confused and were able to adapt our communication and development accordingly. A key outcome has been adoption: our tools now have a dedicated user community that continues to interact with the development team by pointing out bugs and actively guiding ongoing development.
Why did this work?
Implementing FAIR principles is hard, and so too is establishing a decentralized data infrastructure in an existing consortium. In our experience such efforts often enough fail or fizzle out, usually without reports being written. We believe that a crucial aspect to the success of this pilot project has been that we approached the problem both from a social and a technical direction. Understanding needs, finding local champions, addressing concerns, and building excitement around adopting a technical solution is in our view as important as great documentation, simple deployment, and intuitive user experience. With this case report we wanted to give an account of our objectives, the actions we have taken to implement them and what we have learned. We hope that other groups will find our experience useful in navigating similar challenges.
Effective strategies
Beginning with in-person workshops: The two in-person workshops we conducted over the summer of 2024 were essential to quickly understand the practical data challenges ENIGMA-PD faces, and to see how FAIR data tools like Nipoppy and Neuroabgel can help address them. Working together for several days on challenging tasks also helped our teams become familiar and built trust in the potential of our collaboration. These experiences were key to creating the motivation and dedication that later helped us make rapid progress on the projects we described here.
Bringing the right people together: The group that came together, represented a mix of perspectives and roles that were essential to the success of these projects (number of individuals in brackets):
-
Data managers (4). Typically, ECRs with access to and deep understanding of their site’s dataset. Motivated by having experienced the challenge of standardizing data firsthand. Critical to curate and process the datasets and resolve issues in person.
-
Project managers (3): Coordinators with deep knowledge of the challenges in the PD-WG, and of the potential of the neuroinformatics tools. Critical for initial needs assessment, social coordination, and communicating the direction of the projects.
-
Developers (4): Staff and trainee ECRs with expert knowledge of the neuroinformatics tools. Critical to provide support in person, and to quickly address identified bugs.
-
Principal Investigators (4): Faculty responsible for their site in the PD-WG. Motivated by a need for more efficient collaboration processes. Critical for giving the green light for their site to participate and for addressing data governance questions.
A key aspect for local champions was having experienced the data management problems at the core of our projects firsthand. In our experience, this is often truer of ECRs who work closely with the data, therefore focusing on ECRs was a good strategy.
The importance of building and training a community of ECRs: Throughout both efforts, the engagement from ECRs was outstanding. Ten highly motivated ECRs across the four pilot sites took an active role in setting up and maintaining their Neurobagel discovery nodes. Many then independently continued adding more data to their nodes and updated them as new imaging data became available. The project fostered a collaborative environment through regular online meetings and discussions on Slack and Discord, where researchers helped each other navigate technical and logistical hurdles, such as applying for access to high-performance computing resources.
The parallel deployment of Nipoppy and Neurobagel: Beginning with the deployment of Nipoppy at each site provided a standardized framework for data organization and automatic tracking of imaging pipelines. This simplified the later data annotation steps needed for the Neurobagel discovery pilot, which benefits from standardized local data.
Focus on ease of use: The simple, user-friendly interfaces of the Neurobagel tools allowed sites to rapidly annotate their local data after limited training. In fact, we used the “See one, Do one, Teach one” (SODOTO) approach - a teaching model that emphasizes hands-on learning - to encourage ECRs to train each other over the course of the workshops and online support sessions. This approach helped demonstrate early success for participants and build capacity for training. The simple command line interfaces for Nipoppy and for deploying the Neurobagel discovery nodes were important to achieve a rapid adoption of these tools.
Tech support: The ORIGAMI team along with the ENIGMA-PD core team members offered continuous tech support via Discord and online meetings. Going beyond online documentation and investing time in one-on-one sessions turned out to be a big help to ensure timely project progress. We found that starting with synchronous training and support events for several sites at once (workshops and online meetings) and then moving to continued support mainly through asynchronous channels (Discord, GitHub, documentation) worked well for us, as it helped train local experts at each site and created a shared space for sites to help each other. We broadly estimate that after the initial workshops, the overall time spent on tech support and training by the core team was about 60 person-hours for the data standardization project with Nipoppy (with the majority of that time spent on support for BIDS conversion), and about 20 person-hours for the data discovery pilot with Neurobagel (with about equal parts spent on support for data annotation and node deployment).
Clear and transparent goal setting: At the launch of both the ENIGMA-PD wide data processing standardization with Nipoppy, and the cross-site discovery pilot with Neurobagel, we shared with the participating sites a clear list of steps that needed to be taken for each dataset and site. This helped break the tasks into understandable steps and clarified the effort required. We also created a leaderboard that showed the progress of each site and dataset through these steps. The leaderboard served as a nice incentive and reminder to the sites and helped speed up progress.
Lessons learned
Adoption depends on the perceived effort and benefit: From talking to participants, we have learned that their understanding of what was required of them and what the outcome would mean changed quite a bit over the course of both projects and differed from the perspective of the coordinating team. Feedback collected after the project highlights this initial confusion for participants, and the risk this poses to adoption. Even if the tools and processes are easy to use and provide great benefits, unless that is clearly communicated upfront, participants may decide that their limited time is better spent elsewhere. Three factors helped us overcome this challenge during the projects: 1) Participants were familiar with the problem of standardized data processing and discovery ahead of time, likely increasing their willingness to try something new, 2) Participants noticed the support and commitment from the core team, which alleviated some worries about feasibility, 3) Doing in-person workshops made it easy to identify and address the initial confusion, and also meant that participants had committed time to deeply understanding the process. Going forward, we will focus on clarifying the initial investment needed at the beginning of the process and to highlight the expected benefits through case reports and testimonials of successful projects such as this one.
Limitations of standardized vocabularies: Neurobagel depends on existing standardized vocabularies that sites use to annotate their local data. For many data types like demographics, clinical diagnoses, and imaging modalities, excellent vocabularies exist. But the discovery pilot project also highlighted the limitations of even well-curated vocabularies like SNOMED-CT to capture the broad range of clinical assessments captured across sites. Addressing these limitations will likely require a community-led effort to collect and curate a list of published assessment instruments that are commonly used in a neuropsychological context, with links to terms in existing ontologies. A second, compatible solution is to enable a consortium like ENIGMA-PD to create its own internal vocabulary of assessments to use for harmonization among consortia sites. For neuroimaging pipelines, such vocabularies do not exist, so we created our own vocabulary of well-known pipeline names that served as a list of standardized terms (https://github.com/nipoppy/pipeline-catalog). We hope to expand this vocabulary into a community-maintained resource going forward.
Added deployment responsibility to the sites: The unplanned need for sites in this pilot to also handle the deployment of their discovery nodes themselves required additional technical support and highlighted the need to plan for dedicated IT support resources in the future. We think that a good strategy would be to formally approach existing national research compute organizations like SURF in the Netherlands and the Digital Research Alliance in Canada who provided the dedicated cloud infrastructure we relied on in this pilot project. Not only do similar organizations exist in many countries participating in ENIGMA-PD, they also often provide excellent technical documentation, training, and guidance to the researchers in their area of mandate. These resources can help reduce the support burden for future sites looking to deploy and add their own discovery nodes to the network.
But given the need for ongoing maintenance of these services and the technical skillset required to do so, sites need to be able to delegate this task to trained system administrators eventually. Particularly for sites in lower-resourced countries this may create an adoption barrier if no comparable national research compute infrastructure exists or the technical expertise is not locally available. However, we see several mitigation strategies. First, the minimal compute resource needs and simple setup process for a Neurobagel discovery node allow deployment even on very basic hardware as long as local technical expertise is available. Second, even if technical expertise is not locally available, the decentralized nature of Neurobagel allows for more technical sites to act as hubs that host discovery nodes for other sites that retain control of their own nodes. For this, sites only need to agree to move the harmonized data availability records to the node hosted by the hub. Third, if this option is unavailable, sites can still annotate their data to facilitate local introspection of data availability. Even if this does not allow for decentralized discovery of the data, the annotations will still help with the integration of the data once they are eventually transferred to the PD-WG coordinating site in Amsterdam.
Future plans
Broader adoption within ENIGMA-PD: Based on these encouraging results, we plan to continue our collaboration and expand the decentralized data infrastructure of ENIGMA-PD with Nipoppy and Neurobagel. A natural next step would be to invite other sites in the PD-WG who have made good, early progress in adopting the Nipoppy framework to enable upgrades to their image processing pipelines. Since launching the Nipoppy adoption project in December of 2024, 12 sites out of 28 participating in the effort have already completed the setup successfully and are thus in a great position to also make their local data discoverable to the rest of the PD-WG by deploying a Neurobagel discovery node. We will communicate the outcome of this pilot at the next ENIGMA-PD call and invite sites to join.
Improvements of neuroinformatics tools: The second direction of expansion is to extend the neuroinformatics tools to support additional needs of the consortium. For Nipoppy this includes the addition of new pipelines, including: 1) an FSQC (https://github.com/Deep-MI/fsqc) “extractor” that generate quality assurance/quality control metrics and visual summaries on FreeSurfer outputs and 2) a FreeSurfer subsegmentation pipeline to extract subnuclei volumes from subcortical regions. For Neurobagel, the standardized data model can be extended to query additional variables beyond demographics and imaging modalities. Natural candidates here are data types required by recently proposed projects in the PD-WG, for example more advanced clinical variables such as clinical questionnaires for non-motor symptoms, details on symptom onset and side, the availability of genetic data and PET/SPECT data. The flexibility of the graph-based Neurobagel data model will facilitate these expansions.
Governance: A third direction of work will be the creation of a formal, decentralized governance model for the neuroinformatics tools. The purpose of this governance model will be the creation of a community of institutes and stakeholders who will participate in the steering and long-term sustainability of the development, and who will contribute to the necessary training and support for a growing user base.
Expanding adoption beyond ENIGMA-PD: The success of this pilot project has helped us gain a better sense of the feasibility of adopting a decentralized, FAIR data infrastructure for a clinical research consortium. We believe that an initial deployment at the Dutch sites will be an ideal demonstration that will build the case for a subsequent adoption also among international ENIGMA-PD sites. But the challenges we have begun addressing in this pilot project are also common for other ENIGMA working groups and other clinical research consortia in general. Recently, we have agreed to help a newly started ENIGMA working group on tremor that is also led from Amsterdam UMC to adopt the same data processing and discovery technology with Nipoppy and Neurobagel. Additionally, the Netherlands hosts several other ENIGMA working groups (including Autism, Anxiety, Obsessive-Compulsive Disorder, EEG), creating further potential for cross-domain extensibility and long-term sustainability of these efforts. We believe that the strategy of addressing social and technical challenges together is a promising approach to drive the adoption of more efficient, FAIR workflows also for other clinical research consortia. Our goal is to help such interested groups, especially within ENIGMA to adopt this decentralized and modular approach to more standards-based collaboration. We will publish future updates on the progress of this project on this website: https://enigma-infra.github.io/.
Conclusion
The objective of our pilot project was to make four large, internationally distributed datasets of the ENIGMA Parkinson’s Disease working group publicly discoverable and interoperable. By combining the strong social infrastructure of the PD-WG with lightweight neuroinformatics tools for FAIR data processing and discovery, we were able to successfully achieve this objective in a short time frame. As a result of this pilot, for the first time, there is now a discovery portal where the locally available data can be queried live, both by the PD-WG and by the wider public.
This findability across the decentralized, access-controlled data addresses an important need of the PD-WG for determining the feasibility of new research projects, and for tracking sites’ progress towards processing data for ongoing projects. It replaces a time-consuming effort of sending manual data availability forms that had become a bottleneck for efficiency of the PD-WG. The discovery portal is already proving useful for the pilot sites and creates new possibilities for collaboration and integration with researchers and consortia beyond the consortium.
In the process of completing this pilot, we have learned valuable lessons about successful strategies and remaining challenges that have informed our plans to continue the rollout of these technologies to additional international PD-WG sites in the future. Our vision is that, by successfully establishing this decentralized neuroinformatics infrastructure built on open science principles within ENIGMA-PD, we will provide for our community a modern, scalable model for collaborative neuroscience research. We believe that these efforts can enable us to practice and promote open science despite evolving privacy regulations and adapt to paradigm shifts in research setups of the future.
Data and Code Availability
The software that was developed and used in the course of this project is publicly available with open source licenses under the Neurobagel (https://github.com/neurobagel) and Nipoppy (https://github.com/nipoppy/) organizations on GitHub.
Individual ENIGMA-PD sites retain ownership of their MRI scans and only share the anonymized and derived data for analysis. Data are thus not openly available, but researchers are invited to join the ENIGMA-PD Working Group where they can formally request derived data via secondary proposals. Data requests are then considered by the individual site’s principal investigators. If you are interested in joining ENIGMA-PD, please contact enigma-pd@amsterdamumc.nl. For more information, please see the working group website: https://enigma.ini.usc.edu/ongoing/enigma-parkinsons/. The data discovery portal discussed in this article is publicly available at https://enigma.neurobagel.org/.
Funding Sources
Sebastian Urchs: Tanenbaum Open Science Institute; Nikhil Bhagwat: Tanenbaum Open Science Institute; Mathieu Dugré: NSERC, Concordia University; Michelle Wang: CIHR, UNIQUE, Brain Canada, FRQS, Parkinson Québec, FRQNT; Rick C. Helmich: Netherlands organization for Scientific Research (VIDI grant # 09150172010044), Joint Programme Neurodegenerative Disease Research (grant # JPND2024-280); Chris Vriend: MJFF-022801; Ysbrand D. van der Werf: NIH R01NS136995, NIH R01AG058854-03, NIH 1RO1NS107513-01A1, MJFF-021683; Jean-Baptiste Poline: NIH-NIBIB P41 EB019936, CIHR PJT-185948, NIH-NIMH R01 MH083320, NIH RF1 MH120021, NIMH R01MH096906, CZI EOSS5-0000000401, Brain Canada.
Conflicts of Interest
All authors declare no conflicts of interest.