Introduction
The OHBM Brainhack is a community-driven satellite event of the Organization for Human Brain Mapping (OHBM) annual meeting, designed to foster collaboration, innovation, and inclusivity in neuroimaging research. It brings together scientists of diverse backgrounds and career stages to work on open projects, learn new skills, and build lasting professional networks. Central to Brainhack’s mission is the promotion of open science principles—transparency, reproducibility, and accessibility—through hands-on learning and collective problem-solving.
Each year, Brainhack evolves to meet the needs of its growing and increasingly diverse community. The 2023 edition continued this tradition by focussing on three core pillars: 1) Inclusivity – ensuring all participants, regardless of background or experience, feel welcome and empowered to contribute; 2) Empowerment – offering educational programme that supports both structured and self-directed learning; 3) Innovation – encouraging bold, interdisciplinary thinking through collaborative project development. These guiding principles shaped both long-standing and newly introduced components of the 2023 Brainhack.
To support these goals, Brainhack 2023 featured several key components. The Train-Track offered hands-on educational sessions, tutorials, and self-directed learning opportunities tailored to various skill levels and research interests. Meanwhile, the long-standing Hack-Track remained at the core of the event, providing a space for collaborative project work spanning topics such as neuroimaging software, data standards, visualisation, and reproducibility (reported at Section 6). The Buddy System, first introduced in 2022 and refined this year, paired newcomers with experienced attendees to foster engagement, peer support, and a sense of community.
The hybrid format, introduced in recent years, was retained in 2023 to increase accessibility and reduce the event’s environmental impact.1 This approach enabled participation from a broader global audience and addressed challenges such as limited funding and visa restrictions, furthering Brainhack’s commitment to equitable access and global inclusion. This year also saw the introduction of new community engagement initiatives, adding a playful element that enhanced participant connection and involvement. These included both a Mini-Grant initiative to support values-aligned projects and a Rhyming Battle to foster community engagement.
In this manuscript, we offer a structured overview of the OHBM Brainhack 2023 by outlining its planning, hybrid logistics, and community values. We then describe the core components of the event—Train-Track, Hack-Track, and Buddy System—followed by newly introduced initiatives, the Mini-Grant initiative and Rhyming Battle. We also report findings from a post-event participant survey and share lessons learned. This account aims to inform future Brainhack events and inspire continued growth within the open neuroscience community.
Train-Track
Andrea Gondová
The Train-Track was an educational component aiming to create a supportive environment for knowledge exchange, allowing participants to enhance their technical skills and knowledge through both structured and flexible learning sessions. Suggested training materials covered diverse range of topics, including coding skills, good coding and best research practices, and machine learning. These materials consisted of tutorials and/or pre-recorded videos, largely sourced from the BrainHack School (for more information, see Table 1 for training materials and links). Additionally, participants were encouraged to customize their learning experience by self-organising study group with others sharing similar learning goals. On-site sign-ups were also available for mentor-mentee pairings, offering attendees opportunities to gain expertise in advanced topics while expanding their professional networking. Various communication platforms, including in-person sessions and Discord, were used to facilitate collaborative learning.
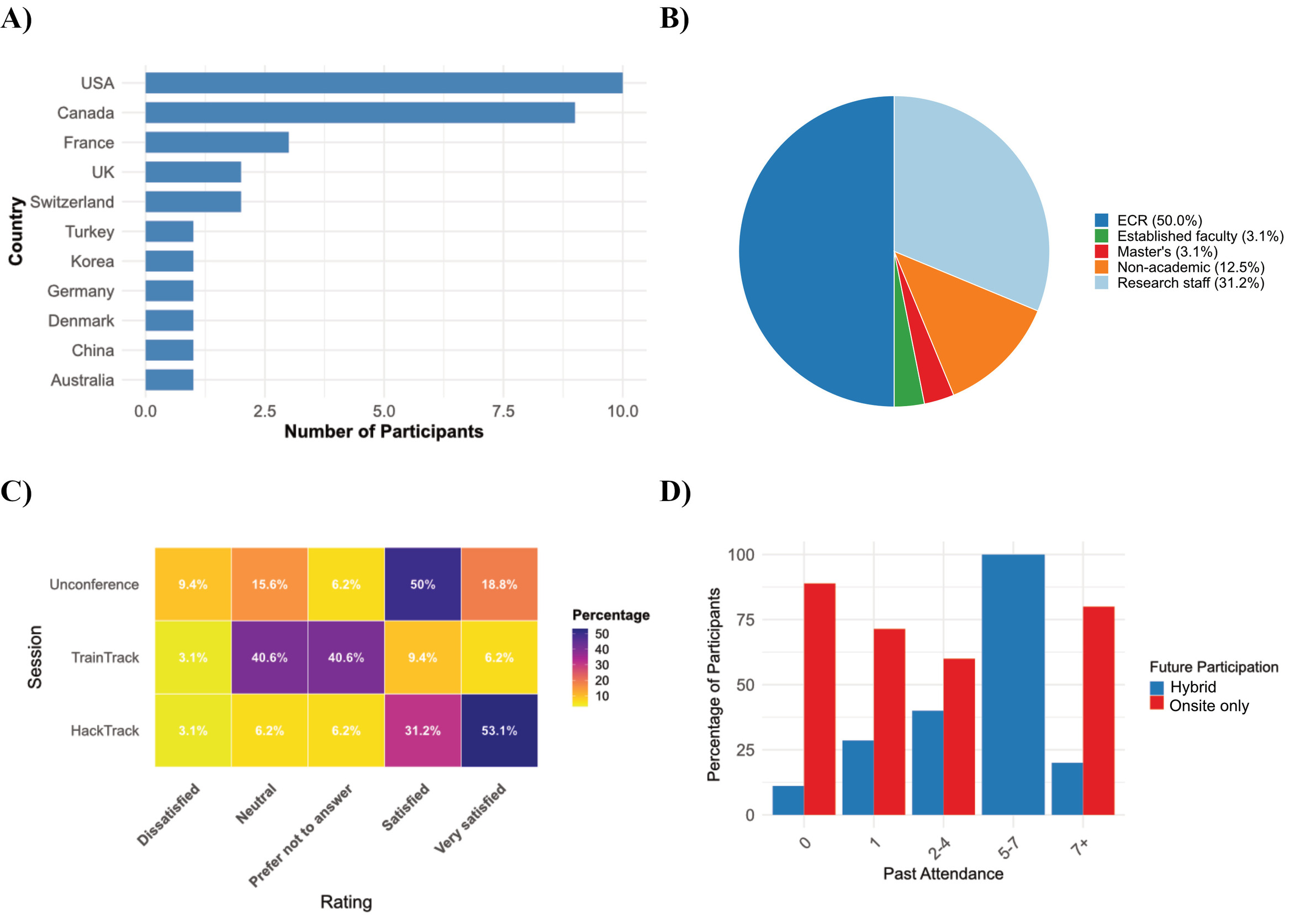
Among the 31 respondents to the post-Hackathon survey, 23% participated in Train-Track sessions, with topics ranging from coding and machine learning to version control and coding best practices. “Good practices” was the most recommended topic by participants. However, feedback suggested a need for more structure and planning, such as scheduled sessions and topic prompts, to improve accessibility and participation. Organisers acknowledged the value of increased structure while maintaining Train-Track’s dynamic and community-driven nature. The goal remains to provide a flexible learning environment with suggested materials for inspiration, rather than prescriptive guidelines. To improve future iterations of Train-Track, organisers plan to introduce the program more clearly at the start of the event, encourage early interest in specific topics, and refine the program’s structure and objectives to better support participants.
Hack-Track
Qing (Vincent) Wang
The Hack-Track is designed for hands-on project development, allowing participants to collaborate on diverse and innovative projects. Before the OHBM Hackathon 2023 began, project leaders submitted their proposals to our repository on GitHub, setting the stage for a dynamic and inclusive event. A pre-Hackathon online session advertised the schedule and introduced participants to the online platform. At the start of the Brainhack, project leaders pitched their projects, outlining goals, current status, and future aspirations.
The projects covered a wide range of topics, including coding, research tool development, documentation, community guideline development, and visualisation techniques. This variety allowed participants to choose projects based on their interests and goals. Some focused deeply on a single project, while others divided their time among multiple projects for a broader experience. Participants selected projects to contribute to their expertise or to acquire new skills and knowledge. Hackathon projects often introduced novel methods or programming languages (e.g., Julia, WebAssembly, new visualisations), providing participants with practical skills for future endeavours. Beyond the technical and educational aspects, the event also fostered community building, with many participants drawn to projects for the opportunity to collaborate and learn from new peers. Over three intensive days, 33 projects advanced significantly, thanks to the collaborative efforts of 167 scientists from across the globe.
Despite the challenges of a hybrid event, especially in synchronizing video streams, we utilised various technologies within Discord to facilitate seamless participant exchanges. To support this, a dedicated Discord server was structured with general-purpose channels (e.g., for questions, announcements, and unconference topics) and separate project-specific channels, allowing teams to share files, code, and updates continuously. During live sessions, such as the unconferences, on-site presentations were streamed directly through Discord, allowing remote participants to watch, ask questions, and contribute in real time. This structure ensured equal access to information, spontaneous collaboration, and dialogue between locations throughout the event. A screenshot illustrating the full channel layout is provided in Figure 2.
Buddy System
Sina Mansour L.
The Buddy System was established to support and engage newcomers at the Hackathon. This initiative pairs participants in a mentor-mentee fashion, with mentors guiding interests, projects to join, and networking opportunities throughout the Hackathon. Introduced in the previous 2022 edition, the system was refined for 2023 to create a more dynamic and natural community structure (see Figure 3).
Pairs were given the freedom to choose their buddies to avoid any biases in matching, ensuring a fair and supportive environment for all participants. In total, 27 mentors and 69 mentees signed up to participate. In doing so, first-time attendees and more experienced brainhackers were encouraged to network and get to know each other while also sharing information about what to expect from the Brainhack. This system has been instrumental in helping first-time attendees feel welcomed and integrated into the community.
Public Relations and Outreach
Bruno Hebling Vieira, Xinhui Li
Following the successful engagement of the OHBM Brainhack 2022, prior to the Hackathon, we leveraged various social media platforms, including Twitter, Mattermost, and WeChat, to announce registration information and promote community engagement by sharing live project presentations during the event. Throughout the Hackathon, Discord served as the main communication platform between organisers and participants, with different channels dedicated to specific purposes. A general announcements channel, first created for Brainhack 2022, was reused in 2023, and many project-specific channels were continued without needing to be recreated. Overall, digital outreach remains a fundamental tool for dissemination, with approximately 30% of the pre-Hackathon survey respondents (N=167) reporting that they learned about the event through social media or the website.
Projects
BrainViewer: Seamless Brain Mesh Visualisation with WebGL
Authors: Florian Rupprecht, Reinder Vos de Wael
BrainViewer, a novel JavaScript package, is poised to transform how researchers and developers interact with brain meshes by enabling seamless visualisation directly within web browsers. By harnessing the power of ThreeJS, BrainViewer offers a responsive and adaptable viewing experience. Its integration with the JavaScript event system facilitates the creation of intricate and customizable behaviours, enhancing its utility.
BrainViewer is currently in an active phase of development. With our commitment to meeting the demands of our short-term projects, we anticipate the first release of BrainViewer within the coming months. BrainViewer is available at Github. Upon release, it will be installable through the npm package management system. A live demo that moves based on microphone input is available at GitHub.
BrainViewer harnesses the power of WebGL and WebGPU (depending on device capabilities) to efficiently render brain meshes within web browsers, delivering a visually engaging experience. Test users have consistently praised the smoothness of interaction. It supports a wide range of devices, from large PC screens with mouse and keyboard to compact mobile screens with touch interfaces, ensuring seamless deployment across ecosystems. Moreover, BrainViewer integrates with the JavaScript event system, enabling users to define complex and tailored behaviours that enhance interactivity and exploration.
Clinica’s image processing pipeline QC
Authors: Matthieu Joulot, Ju-Chi Yu
The goal of the project was to evaluate different quality check (QC) metrics and visuals for pipelines currently existing in Clinica,2 which deal with registration or segmentation. That way, we would be able to find a few good metrics to separate the good images from the bad or moderately good ones, which the users would check using some visual we would generate for them.
We were able to output seven available QC metrics for registration, and with PCA, we evaluated their factor structure and kept only two metrics that capture the most QC information: 1) the correlation coefficients between the MNI template (target) and the subject’s brain image, and 2) the Dice score (i.e., a type of distance measure) obtained by a brain extraction tool (BET) algorithm called HD-BET 4.
We will look further into our pipeline based on this result, so that we can have a better idea of what we want to implement in the future version of Clinica, which will include QC.
Neuroimaging Meta-Analyses
Authors: James Kent, Yifan Yu, Max Korbmacher, Bernd Taschler, Lea Waller, Kendra Oudyk
Neuroimaging Meta-Analyses serve an important role in cognitive neuroscience (and beyond) to create consensus and generate new hypotheses. However, tools for neuroimaging meta-analyses only implement a small selection of analytical options, pigeon-holing researchers to particular analytical choices. Additionally, many niche tools are created and abandoned as the graduate student who was working on the project graduated and moved on. Neurosynth-Compose/NiMARE are part of a Python ecosystem that provides a wide range of analytical options (with reasonable defaults), so that researchers can make analytical choices based on their research questions, not the tool.
To enhance and broaden the ecosystem, our efforts were channelled into a variety of projects. Initially, we focused on making Coordinate-based Meta-Regression more efficient and user-friendly, with the goal of fostering the adoption of a more flexible and sensitive model approach to coordinate-based meta-analysis. Following that, we sought to improve the tutorial for Neurosynth-Compose, aiming to increase its usage by providing clear and accessible guidance on how to utilise the website effectively. Additionally, we aimed to refine the masking process for Image-Based Meta-Analysis, striving to incorporate more voxels/data, thereby enhancing the analysis’s overall data richness. Finally, we embarked on running topic modelling of abstracts from papers associated with NeuroVault collections, a move intended to identify groupings of images that are suitable for meta-analysis, further contributing to the field’s advancement and the utility of NeuroVault’s resources.
Progress was achieved across all the projects undertaken to enhance the ecosystem. For the Coordinate-based Meta-Regression, we identified several bugs and areas of inefficient code, alongside creating notebooks that demonstrate usage and pinpoint issues, laying the groundwork for future improvements. Feedback was meticulously provided to refine the tutorial, focussing on improving its clarity and conciseness, thereby making it more accessible to users. In terms of the Image Based Meta-Analysis, an outline of a solution to include more voxels was drafted, complete with a plan for its implementation, promising to enrich the analysis by utilizing a larger dataset. Furthermore, through topic modelling, we gained insights into the distribution of images on NeuroVault, identifying how they are grouped, which aids in understanding the landscape of available data for meta-analysis.
The improvements made to NiMARE and related tools provide more accessibility to neuroimaging meta-analyses making it easier to perform crucial analyses in our field.
NARPS Open Pipelines
Authors: Boris Clénet, Élodie Germani, Arshitha Basavaraj, Remi Gau, Yaroslav Halchenko, Paul Taylor, Camille Maumet
Different analytical choices can lead to variations in the results, a phenomenon that was illustrated in neuroimaging by the NARPS project.3 In NARPS, 70 teams were tasked to analyse the same dataset to answer nine yes/no research questions. Each team shared their final results and a COBIDAS-compliant4 textual description of their analysis. The goal of NARPS Open Pipelines is to create a codebase reproducing the 70 pipelines of the NARPS project and share this as an open resource for the community.
During the OHBM Brainhack 2023, participants had the opportunity to improve the project in various ways. First, they focused on fostering a more inclusive environment for new contributors, refining the contribution process through proofreading and testing. Several pull requests (PR #66, PR #65, PR #63, PR #64, PR #52, PR #50) were dedicated to this effort, rectifying inconsistencies in the documentation and contributing procedures. Additionally, they implemented GitHub Actions workflows to enable continuous integration, ensuring the testing of pipelines with each alteration (PR #47), and crafted a new workflow to detect code comments and documentation typos (PR #48). Acquisition of new skills was another significant aspect, with individuals diving into NiPype and advancing pipeline reproductions. Thanks to Paul Taylor’s contributions, a deeper understanding of AFNI pipelines emerged, particularly benefiting from afni_proc examples within the NARPS project.5 This endeavour resulted in the initiation of new pipeline reproductions (PR #62, Issue #61, PR #59, Issue #57, Issue #60, PR #55, Issue #51, Issue #49). Ultimately, the Hackathon yielded substantial progress: 6 pull requests merged, 4 issues opened, 4 closed, and 6 opened, showcasing a fruitful collaboration and dedication to project enhancement.
ET2BIDS - EyeTracker to BIDS
Authors: Elodie Savary, Remi Gau, Oscar Esteban
In the realm of neuroimaging, BIDS has become the de facto standard for structuring data. Its adoption has simplified the data management process, enabling researchers to focus on their scientific inquiries rather than the intricacies of data organisation.6 However, the inclusion of eye-tracking data within the BIDS framework has remained a challenge, often requiring manual data rearrangement. The ET2BIDS project aimed to take a significant step toward incorporating eye-tracking data into BIDS, leveraging the bidsphysio library to facilitate this integration.
Bidsphysio is a versatile tool designed to convert various physiological data types, including CMRR, AcqKnowledge, and Siemens PMU, into BIDS-compatible formats. In particular, it offers a dedicated module called edf2bidsphysio to convert EDF files containing data from an Eyelink eyetracker. An advantageous feature of the bidsphysio library is its Docker compatibility, ensuring smooth cross-platform execution without the need for additional installations.
Before starting our project, a bug had been identified in the Docker image, which hindered the correct execution of the edf2bidsphysio module. Our primary focus was, therefore, to address and resolve this bug in order to enable the utilisation of the Docker image for eye-tracking data conversion.
The ET2BIDS project made significant progress in converting eye-tracking data into BIDS format. We initially processed the test data from the GitHub repository, overcoming the bug that had hindered the edf2bidsphysio module’s execution. Moreover, with minor modifications to the module, we successfully processed a dataset acquired by the authors using an Eyelink eye tracker, showcasing potential bidsphysio versatility in handling various eye-tracking datasets.
The future of eye-tracking data integration into BIDS is evolving as we identify essential fields and metadata necessary for reproducible research. BIDS specifications for eye-tracking data are in development, with expanding guidelines for essential reporting in research studies (e.g. 7).
As these guidelines grow, we will have to adapt bidsphysio to match the evolving BIDS standards, ensuring it converts eye-tracking data in accordance with the latest recommendations.
Physiopy - Practices and Tools for Physiological Signals
Authors: Mary Miedema, Céline Provins, Sarah Goodale, Simon R. Steinkamp, Stefano Moia
The acquisition and analysis of peripheral signals such as cardiac and respiratory measures alongside neuroimaging data provides crucial insight into physiological sources of variance in functional magnetic resonance imaging (MRI) data. The physiopy community aims to develop and support a comprehensive suite of resources for researchers to integrate physiological data collection and analysis with their studies. This is achieved through regular discussion and documentation of community practices alongside the active development of open-source toolboxes for reproducibly working with physiological signals. At the OHBM 2023 Brainhack, we advanced physiopy’s goals through three parallel projects:
Documentation of Physiological Signal Acquisition Community Practices: We have been working to build “best community practices” documentation from experts in the physiological monitoring realm of neuroimaging. The aim of this project was to draft a new version of our documentation adding information from six meetings throughout the year discussing good practices in acquisition and use of cardiac, respiratory, and blood gas data. The documentation is finished and ready for editorial review from the community before we release this version publicly.
Semi-Automated Workflows for Physiological Signals: The aim of this project was to upgrade the existing code base for the peakdet and phys2denoise packages to achieve a unified workflow encompassing all steps in physiological data processing and model estimation. We mapped out and began implementing a restructured workflow for both toolboxes incorporating configuration files for more flexible and reproducible usage. To better interface with non-physiopy workflows, we added support for NeuroKit2 functionalities. As well, we added visualisation support to the phys2denoise toolbox.
PhysioQC: A Physiological Data Quality Control Toolbox: This project was about creating a quality control pipeline for physiological data, similar to MRIQC, and leveraging NiReports. At the Hackathon, we implemented a set of useful metrics and visualisations, as well as a proof-of-concept workflow for gathering the latter in an HTML report. Going forward, development of these toolboxes and revision of our community practices continues. We welcome further contributions and contributors, at any skill level or with any background experience with physiological data.
Cerebro - Advancing Openness and Reproducibility in Neuroimaging Visualisation
Authors: Sina Mansour L., Annie G. Bryant, Natasha Clarke, Niousha Dehestani, Jayson Jeganathan, Tristan Kuehn, Jason Kai, Darin Erat Sleiter, Maria Di Biase, Caio Seguin, B.T. Thomas Yeo, Andrew Zalesky
Openness and transparency have emerged as indispensable virtues that pave the way for high-quality, reproducible research, fostering a culture of trust and collaboration within the scientific community.8 In the domain of neuroimaging research, these principles have led to the creation of guidelines and best practices, establishing a platform that champions openness and reproducibility.9 The field has made substantial strides in embracing open science, a feat achieved through the successful integration of open-source software,10 public dataset sharing,11 standardised analysis pipelines,4 unified brain imaging data structures,6 open code sharing,12 and the endorsement of open publishing.
The joint efforts of these initiatives have established a robust foundation for cultivating a culture of collaboration and accountability within the neuroimaging community. Nonetheless, amidst these advancements, a crucial inquiry arises: What about the visual representations —the very tools through which we observe and make sense of neuroimaging results? It is within this context that we introduce Cerebro,13 a new Python-based software designed to create publication-quality brain visualisations (see Figure 5). Cerebro represents the next step in the quest for openness and reproducibility, where authors can share not only their code and data but also their publication figures alongside the precise scripts used to generate them. In doing so, Cerebro empowers researchers to make their visualisations fully reproducible, bridging the crucial gap between data, analysis, and representation.
As Cerebro continues to evolve in its early development stages, this manuscript serves as a manifesto, articulating our dedication to advancing the cause of open and reproducible neuroimaging visualisation. Cerebro is guided by a set of overarching goals, which include:
Fully Scriptable Publication-Quality Neuroimaging Visualisation: Cerebro’s primary mission is to equip researchers with the means to craft impeccable brain visualisations while preserving full scriptability. Cerebro enables researchers to document and share every aspect of the visualisation process, ensuring the seamless reproducibility of neuroimaging figures.
Cross-Compatibility with Different Data Formats: The diversity of brain imaging data formats can pose a significant challenge for visualisation. Many existing tools are constrained by compatibility limitations with specific formats. Drawing upon the foundations laid by tools like NiBabel,14 Cerebro is determined to provide robust cross-compatibility across a wide spectrum of data formats.
Integration with Open Science Neuroimaging Tools: We wholeheartedly acknowledge that Cerebro cannot thrive in isolation. To realize its full potential, it must seamlessly integrate into the existing tapestry of open science tools, software, and standards. Through active collaboration with the neuroimaging community, particularly through open, inclusive community initiatives such as brainhacks,15 Cerebro aspires to forge cross-compatibility with established and emerging tools, standards, and pipelines. Our vision is one where the future of neuroimaging visualisations is marked by unwavering openness and reproducibility, fostered by this united effort.
Publishing Code in Aperture Neuro
Authors: Renzo Huber, M. Mallar Chakravarty, Peter Bandettini
In the dynamic field of neuroimaging, researchers derive new insights into the brain through the application and development of innovative software and analysis tools. While scientific findings can be described in conventional PDF papers, many other aspects of researchers’ work often do not go through a similar publication procedure, lacking peer review and full citability. These research outputs hold significant value for the neuroimaging community. Examples include code, code wrappers, pipelines, toolboxes, toolbox plugins, and code notebooks.
Although these research objects are typically not subject to peer review and are not indexed in standardised publication systems, the quality of research outputs is directly dependent on the quality of these tools. Hence, it is critical that these objects are citable in a manner that recognizes the author’s contribution, aids in the assessment of scholarly impact, such as their h-index, and promotes a dynamic and reproducible research enterprise. At the time of the Brainhack in July 2023, the OHBM journal Aperture Neuro aimed to broaden its range of submission formats, surpassing the constraints of traditional PDFs, by incorporating code submissions. In a special initiative, Aperture Neuro invited members of the neuroimaging community to submit their research objects in the form of code. The purpose of the Brainhack 2023 project was to discuss this initiative with members of the OHBM coding community and reach a community consensus on the most suitable publication mechanisms. As part of this Brainhack project:
-
We compiled a list of the most impactful, previously unpublished code;
-
We defined the focus of Aperture Neuro’s code submissions with respect to openness, novelty, utility to the community, scientific need, coding style, reproducible executability, and other relevant criteria;
-
We evaluated the trade-offs between different approaches to representing evolving research outputs: the timeless DOI-linked printed document versus dynamic, continuously developing code.
Ever since the Brainhack project in July 2023, Aperture Neuro has launched a call for submissions at https://apertureneuro.org/pages/533-code. Multiple code objects have been published already. Under this publication mechanism, each code submission undergoes assessment by an expert, and when accepted, is aimed to receive a PubMed-listed index, DOI, and a citable reference.
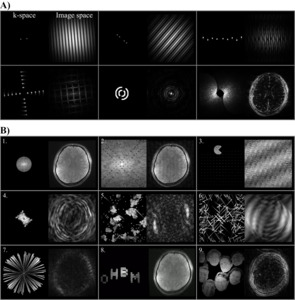
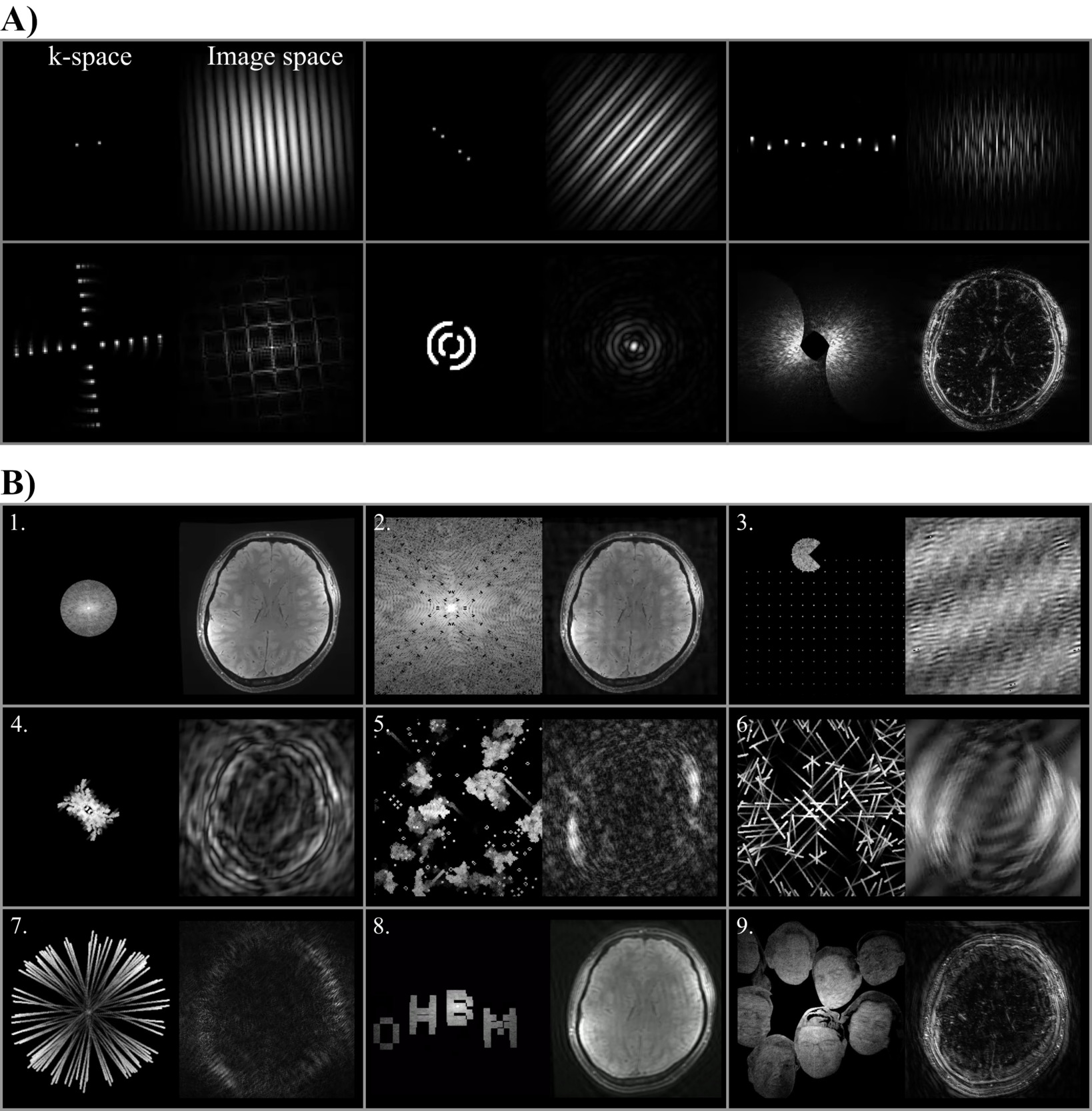
K-Particles: A visual journey into the heart of magnetic resonance imaging
Authors: Omer Faruk Gulban, Kenshu Koiso, Thomas Maullin-Sapey, Jeff Mentch, Alessandra Pizzuti, Fernanda Ponce, Kevin R. Sitek, Paul A. Taylor
The primary objective of this project is to leverage particle animations to provide insights into the journey of traveling and sampling the k-space. By generating captivating visualisations, we aim to demystify the concept of k-space and the role it plays in MRI, engaging both the scientific community and the general public in a delightful exploration of this fundamental aspect of imaging. This project builds on our previous Brainhack experiences that focused on generating 3D geodesic distance computation animations16 and particle simulation-based brain explosions.17
Our methodology is as follows. First, we start from a 2D brain image (e.g. selecting a slice from a 3D anatomical MRI data.18–20 Then, we take its Fourier Transform and subsequently perform masking (or magnitude scaling) operations on k-space data,21 the transform. We then create simultaneous visualisations of the k-space magnitude and corresponding image-space magnitude data. Note that the masking of k-space data is where the participants of this project exercised their creativity, by setting up various initial conditions and a set of rules to animate the mask data. In the final step, animated frames are compiled into movies to inform (and entertain). The scripts we have used to program these steps are available in GitHub. Note that, we have also included several animations where no brain images were used, but instead, we generated k-space data directly in k-space to guide the unfamiliar participants with the concepts (see Figure 7, panel A).
As a result of this Hackathon project,a compilation of our progress (see Figure 7, panel B) can be accessed via YouTube as a video. Some of the highlights are:
-
Audio-visualiser that maps audio features (e.g. amplitude) to k-space mask diameters.
-
Game of life22 simulation in k-space with Hermitian symmetry.
-
Pacman moving in k-space is implemented as a series of radial sector masks.
-
Semi-random initialization of particle positions and semi-randomly reassigned velocities that look like an emerging butterfly.
-
Randomized game of life initialization with vanishing trails of the previous simulation steps that look like stars, clouds, and comets orbiting frequency bands in k-space.
-
Semi-random initialization of particle positions and velocities that look like explosions.
-
Predetermined initialization of particle positions (e.g. at the center) and semi-randomized velocities that look like fireworks.
-
Dancing text animations where the positions of text pixels are manipulated by wave functions.
-
Picture based (e.g. cropped faces of the authors) moving within the k space where each pixel’s grayscale value is mapped onto a mask coefficient between 0-1.
Our future efforts will involve sophisticating the k-space simulations to generate more entertaining and educational content. For instance, instead of only visualising the magnitude images, we can generate four panel animations showing real and imaginary (or magnitude and phase) components of the data.
Mini-Grant Initiative
Yu-Fang Yang and Anibal Sólon Heinsfeld
This year, we introduced Mini-Grant initiative, generously supported by sponsors, aimed at empowering and supporting Brainhackers. The grants were distributed across nine categories:
-
Open Science Mini-Grant: Funding projects promoting open science principles like data sharing and open-source software.
-
Diversity and Inclusion Mini-Grant: Supporting initiatives that enhance diversity and inclusion in science or academia.
-
Public Science Communication Mini-Grant: Backing efforts to communicate scientific research to the public in an accessible manner.
-
Innovative Teaching Method Mini-Grant: Encouraging creative approaches to science education.
-
Interdisciplinary Collaboration Mini-Grant: Fostering cross-disciplinary research to address complex challenges.
-
Student-Led Research Mini-Grant: Supporting science projects led by students.
-
Global Science Collaboration Mini-Grant: Encouraging international research collaborations.
-
Early Career Researcher Mini-Grant: Funding early-career scientists to conduct independent projects.
-
Science and Art Integration Mini-Grant: Funding projects that blend scientific concepts with artistic expression.
These grants were based on the merit of the project, with nominations made by teams or peers after the project pitches and the rhyming battle. The Mini-Grant Initiative was introduced to recognize and support projects aligned with open science, diversity, and interdisciplinary collaboration. Although the initiative was well-intentioned, participation was notably limited. Across the eight grant categories, only nine individuals were nominated in total, with several categories receiving no nominations at all and none exceeding two nominations. When the organising team invited the community to vote among the nominees, there was no meaningful engagement—no participants expressed interest in casting a vote. These outcomes suggest that the competitive format of the initiative may not have resonated with the collaborative ethos of the Brainhack community. This experience provided valuable insight, leading us to reconsider the format for future iterations. Moving forward, we plan to initiate the funding process earlier and integrate it into the registration phase, allowing for a more needs-based and inclusive approach to resource distribution.
Rhyming Battle
Yu-Fang Yang and Anibal Sólon Heinsfeld
One of the most memorable and engaging events at the OHBM Brainhack 2023 was the ‘Rhyming Battle.’ Held before the project presentations, this unique event featured three participants who artistically expressed their experiences and challenges encountered during the Hackathon. They crafted short poems about their projects and the various aspects they faced during Brainhack, bringing a touch of light-hearted humour to their scientific endeavours (see the supplementary material for lyrics). The Rhyming Battle provided an entertaining break from the Hackathon’s more scientific aspects. It allowed attendees to resonate with each other’s experiences, celebrating the struggles and triumphs inherent in scientific projects. The event showcased the creative and vibrant spirit of the Brainhack community. Voting for the winner was not straightforward, as the attendees were equally good. The prize was a single home-made 3D sculpture representing the Brainhack 2023 logo, made for the winner of the battle (Figure 8). However, the enthusiasm of the battlers and the public was overwhelming, and we decided to 3D-print the same sculpture for every battle participant. Ultimately, the spirit of the Brainhack community prevailed,15,23 and every participant was celebrated as a winner.
Conclusion
The OHBM Brainhack 2023 exemplified a vibrant fusion of education, innovation, and community spirit. Through initiatives like Train-Track, Hack-Track, the Buddy System, the Rhyming Battle, and the Mini-Grant initiative, the event fostered an environment rich in learning, creativity, and collaboration. These elements, coupled with a commitment to open science and diversity, underscored the gathering’s success in nurturing a dynamic scientific community. Reflecting on the experiences and feedback from this year, Brainhack is poised to evolve, embracing lessons learned to enhance future hackathons. This continuous improvement and deepened community engagement are set to further empower and inspire participants in the years to come.
Acknowledgements
We thank all the survey participants, contributors, and attendees of Brainhack OHBM 2023 for their generous participation and feedback. We are especially grateful to Feilong for his constructive comments during the review process, which helped to improve the clarity and structure of this report. We also thank OHBM, OSSIG, OpenNeuro, NeuroMod, Child Mind Institute, Canadian Open Neuroscience Platform, TReNDS, BIDS Connectivity Project and other community partners for their support of open neuroscience events. Lastly, we acknowledge volunteers and organisers whose commitment to inclusivity, open science, and community building made this hybrid Brainhack possible.
Funding Sources
QW: Shanghai Mental Health Center key project No. 2023zd01; AB: Australian Government Research Training Program Scholarship and Stipend; MMC: Salary support from les Fonds du Recherche Quebec -Sante; BC: Inria (Exploratory action GRASP); EG: Region Bretagne (ARED MAPIS) and Agence Nationale pour la Recherche for the programm of doctoral contracts in artificial intelligence (project ANR-20-THIA-0018); SEG: NIH - F99AG079810-01; JJ: Royal Australia New Zealand College of Psychiatrists (Beverley Raphael scholarship), NHMRC (GNT2013829), and philanthropic support from the Rainbow Foundation. TM-S: NIH grant [R01EB026859]; CM: Region Bretagne (ARED MAPIS), Agence Nationale pour la Recherche for the program of doctoral contracts in artificial intelligence (project ANR-20-THIA-0018) and by Inria (Exploratory action GRASP); SLM: NIH, NICHD (type: F31; HD111139), Role: PI; AN: Google, Morgan Stanley Foundation, NIMH R21 MH118556-01; M-EP: PhD research scholarship from the Fonds de recherche du Québec - Nature et Technologie (FRQNT); CP: Swiss National Science Foundation —SNSF— (#185872; OE); SRS: Novo Nordisk Exploratory Interdisciplinary Synergy grant (ref NNF20OC0064869) and Lundbeck Foundation postdoc grant (ref R402-2022-1411); TW: Intramural Research Program of the National Institutes of Health; KVH: Dutch Research Council (NWO) Vidi Grant (09150171910043).
Conflicts of Interest
OFG is a full-time employee of Brain Innovation (Maastricht, Netherlands).



_front_of_a_buddy_card._b)_back_of_a_buddy_card._buddy_cards_were_distributed_among_all_.png)

_cerebro_leverages_an_extensive_ar.png)

_skull_sculpture_generated_using_midjourneys_generative_models_for_the_main_website._b).png)

_front_of_a_buddy_card._b)_back_of_a_buddy_card._buddy_cards_were_distributed_among_all_.png)

_cerebro_leverages_an_extensive_ar.png)

_skull_sculpture_generated_using_midjourneys_generative_models_for_the_main_website._b).png)