Introduction
The trajectory of scholarly communication is marked by transformative technological shifts. It began with the meticulously hand-copied manuscripts of medieval scholars. This was followed by the advent of the first academic journals in the 17th century.1 In the 19th and early 20th centuries, typewritten manuscripts were circulated via postal services. By the late 20th century, the digital age ushered in online platforms that enabled widespread electronic dissemination and archival of research articles. While these developments dramatically increased accessibility, they did not fundamentally alter the structure or format of scientific publishing. Most articles remained static PDFs, detached from the underlying data, code, and computational environments that produced their findings.
Today, open-source technologies and reproducibility-driven initiatives create an opportunity to reimagine scientific articles as living documents, encompassing qualities such as interactivity, executability, and true computational depth. This review situates that opportunity within a broader technical landscape. It argues that scholarly publishing is on the brink of a transformation, one that could re-align publication with the ethos of open science, computational reproducibility, and collaborative progress.
The review proceeds in three movements:
-
A diagnostic analysis, exposing the technical limitations and economic asymmetries of the legacy publishing complex. We trace how the infrastructure meant to serve science has instead come to extract disproportionate value from it, and how this dynamic has delayed the adoption of more open, interactive modalities.
-
A survey of enabling technologies, charting the open-source tools and cloud platforms that allow authors to create interactive, executable “living publications.” We highlight NeuroLibre as a case study for end-to-end reproducibility at scale. It demonstrates how code, data, and computational environments can be integrated into a cohesive and shareable research object.
-
A procedural outlook, exploring how distributed peer review, workflow modularization, and new incentive models can support a more transparent and dynamic publishing ecosystem. We introduce the concept of woven literature, building upon Knuth’s original notion of literate programming, to describe publications where prose, computation, and verification not only coexist but evolve together as integrated components.
Through comprehensive analysis and practical recommendations, this review provides researchers, developers, and infrastructure providers with a strategic roadmap for creating, curating, and sustaining the next generation of scientific literature. The practical considerations address critical questions that determine implementation success: What are the resource boundaries of browser-based execution environments? How can large datasets be reproducibly integrated without overwhelming readers or computational platforms? When should upstream, resource-intensive workflows transition to lighter, reader-facing phases of analysis? How can emerging tools ensure complete traceability between raw inputs and final results? How do novel reuse patterns become possible through these advanced publishing technologies?
By addressing these questions, this review bridges the gap between theoretical potential and practical implementation of next-generation scholarly communication systems.
When the elm strangles the vine: A broken equation
The role of academic publishers in advancing scientific progress is undeniable. For centuries, researchers and publishers have shared a mutually beneficial relationship. This partnership is symbolically captured in Elsevier’s logo, which features an old man, representing the scientist, harvesting grapes, a metaphor for knowledge, from a vine entwined around an elm tree, which represents the publisher. The accompanying motto, Non Solus (“not alone”), emphasizes the collaborative nature of scientific discovery.2
The symbiosis between the elm tree and the grapevine is multi-layered and has been cultivated since Ancient Greece.3 The elm provides structural support for the vine, while its canopy helps regulate the microclimate, improving grape quality. During droughts, the tree may even share its sap with the vine, helping it survive under adverse conditions. Elsevier’s logo, though a poetic representation of what science expects from publishers, turns out to be botanically inaccurate: without pruning, the depicted elm would reduce grape yield and undermine its potential to support the vine through sap sharing.4
More critically, this symbolism has become practically reversed: what began as collaboration has tilted toward commodification. Instead of publishers sustaining the growth of science, it is now the scientific enterprise at scale that fuels the commercial success of a few dominant publishers. The primary beneficiaries of the advances in research dissemination have been a few dominant publishers, whose profit margins (up to 40-50%) have outpaced even those of the most successful high-tech companies,5 revealing a disproportion between their economic gains and the actual value they add to scholarly communication.
To begin, this discrepancy can be examined through its numerical dimensions: The real cost of hosting a PDF and registering it as scholarly content is estimated to be around $2.71,6 assuming an open-access online journal that publishes around 300 articles annually (e.g., the Journal of Open Source Software, JOSS7). On the other hand, article processing charges (APCs) of a major publisher can reach a staggering $12,000, with an average of $3.3k across nearly 14,000 publications per year.8 Notably, one such publisher has claimed that the internal cost of publishing an open-access article ranges between $30,000 and $40,000.8 This nearly 4,500-fold disparity on average, which rises to as much as 14,500-fold according to some claims, between the actual cost of $2.71 and the brand-inflated cost of making an article publicly accessible does not correspond to a proportional increase in scientific impact.9
If not delivering impact, is this level of spending at least contributing to the evolution of scientific communication? Unfortunately, an equally pressing concern lies in the underutilization of even basic technological advancements. A telling example is the so-called Continuous Article Publishing model (CAP), introduced in the early 2000s. Here, the term continuous refers to the practice of releasing accepted manuscripts online as they are ready, rather than waiting for complete issues to be compiled. Remarkably, some journals are still in the process of transitioning to CAP more than two decades later. This slow adoption underscores the inertia of legacy publishing systems, which struggle to implement even modest procedural changes, let alone the infrastructural transformation needed to support truly modern and integrated forms of scholarly communication.10
As a result of the inertia embedded in legacy publishing systems, digital publication often remains structurally tied to the logic of print. Publications remain static, disconnected from the data, code, and computational environments that would allow them to serve as dynamic components of an evolving research ecosystem. In signal processing terms, the body of knowledge is sampled but never reconstructed: research exists as discrete outputs without the connective infrastructure for synthesis, independent replication, or reuse.
Encased in this digital shell modeled on print-era conventions, academic papers continue to function as isolated artifacts. The literature becomes increasingly bibliographic, where citations serve more as symbolic gestures than as functional links between interoperable knowledge objects. Sticking with the signal processing analogy, CAP is only about increasing the sampling rate of a bibliographic literature, primarily serving to accelerate publishers’ revenue streams, falling far short of the infrastructural leap required for Continuous Science.11
The concept of Continuous Science represents a fundamental modernization of research dissemination. It promotes a truly continuous digital medium in which articles are no longer isolated relics but integrated research objects.10 Returning to the signal processing analogy, this is not merely about increasing the sampling rate. It is about reconstructing a coherent and interoperable knowledge system. Such a system would enhance the signal-to-noise ratio of scientific communication by enabling reproducibility, synthesis, and meaningful connectivity across articles.
Before turning to the elements of modern solutions that can enable true continuity in open and reproducible scientific knowledge, it is worth considering another perspective on legacy publishing in light of recent advances in generative artificial intelligence (GenAI).
Legacy publishing can’t keep its footing in a GenAI world
Even a decade before large language models (LLMs) made their sweeping entrance into scientific prose,12 the oligopolistic nature of the publishing ecosystem had already been described as the most profitable obsolete technology in history.13
Against this backdrop of stagnation, the pace of innovation in AI has been nothing short of astonishing. The second quarter of 2025, just four days before the writing of this manuscript, witnessed the introduction of ARC-AGI-2, a new benchmark in the Abstraction and Reasoning Corpus for Artificial General Intelligence.14 This update was prompted by a rapid rise in top performance on the previous benchmark, which increased from 34% in early 2024 to 88% by the end of the year (for the latest leaderboard see15). For comparison, members of the general public typically score around 77%, while STEM graduates score approximately 98%.
As LLMs approach expert-level proficiency in generating coherent and convincing prose so rapidly that we need to move the goalpost in favor of humans, it becomes increasingly important to ask: what remains of a publication when we remove the prose?16
Even twelve years before the initial public release of advanced AI agents, this challenge had already been articulated by17: “An article about computational results is advertising, not scholarship. The actual scholarship is the full software environment, code and data, that produced the result.” The legacy publishing system is far too outdated to be retrofitted into an ecosystem where papers serve as more than advertisements.
It is only fair to retire the legacy publishing system with a new acronym: BOOMER (Barely Operational and Obsolete Manuscript Evaluation Rituals). Perhaps it is time to move toward something more relevant and forward-looking for the century we are in.
From BOOMER to next-gen publishing
Science is what we understand well enough to explain to a computer. Art is everything else we do. […] People think that science is the art of geniuses, but the actual reality is the opposite, just many people doing things that build on each other, like a wall of mini stones.
- Donald Knuth18
Scientific progress is often portrayed as the product of isolated breakthroughs by exceptional individuals. Yet as Knuth reminds us, its true foundation is incremental and collective, where people do small, explainable contributions that build on each other. The BOOMER paradigm stands in the way of this accumulative process, perpetuating a format in which knowledge is trapped in static, monolithic documents, divorced from the computational processes and materials that generated it.
Moving beyond this paradigm first requires a return to a more foundational principle that science is what we understand well enough to explain to a computer. This is not merely a technical assertion. It is a claim about findability, accessibility, interoperability, and reproducibility, i.e., the FAIRness of ideas.19 It is also the central premise of Knuth’s concept of literate programming,20 in which the construction of a program is guided not just by what it does, but by how clearly its purpose and logic can be conveyed to others.
Literate programming proposes that narrative and computation should not simply coexist, but be intricately woven together20. It allows a program to function both as a computational artifact and as a document that communicates scientific intent. Even though it has not been proven effective for professional software engineering,21 it has become one of the most preferred approaches for end users who code as part of scientific analysis, also known as end-user software engineering.22 For example, a neuroimaging researcher using literate programming tools, such as Jupyter Notebooks,23 to preprocess MRI data is engaging in end-user programming. In the 21st century where computation has become an indispensable part of almost any scientific discipline, literate programming tools boost exploratory analyses by binding prose, code, data, inputs, and outputs into a single space of reasoning.24 Electronic lab notebooks offer a powerful example of such documentation, capturing the narrative of a researcher’s data exploration to serve as a valuable asset for reproducibility.25
This approach to reproducible documentation naturally informs the design of a modern and federated publishing network.26 Drawing inspiration from Knuth’s concept of literate programming, we can envision woven literature as the natural outcome of such networked scientific publishing. In woven literature, narrative, data, code, and computational environments are stitched together from the outset, not retrofitted after publication. Each thread preserves its own integrity, yet contributes to a fabric that is more than the sum of its parts.
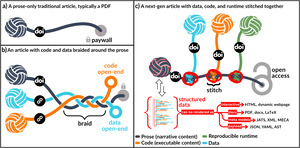
This shift is illustrated in Figure 1. A traditional static article (Figure 1a) presents a self-contained and static document, with a narrative frozen at the time of publication. While it includes references and a DOI that link it to the broader literature, it lacks open and embedded access to the data, code, or computational context that produced its findings. Even when such artifacts are available (Figure 1b), they are typically hosted externally and linked as afterthoughts, which 20% of the time leads to reference rot.27 A next-gen article (Figure 1c), by contrast, embodies the principles of literate programming. It stitches together code, data, runtime, and narrative into a cohesive structure. Moreover, representing the article’s content as structured data enables seamless transclusion of its components into other documents across different platforms. This way, the article becomes not just a description of research but a functional expression of it.
Figure 2 shows how this transformation scales beyond individual articles to the scholarly ecosystem. Bibliographic literature (Figure 2a) supports credit and citation but not actionable reuse. Even when reproducibility artifacts are linked (Figure 2b), they remain external and structurally isolated. Woven literature (Figure 2c) introduces formal, machine-readable relationships among the elements of research.
The conceptualized relationships in Figure 2c, such as reuse data or transclude a figure, unlock several reuse patterns: i) transclusion, when one article directly inserts content from another article such as a paragraph or a figure, ii) longitudinal extension, when a living article re-executes itself with new data points to extend its findings, iii) horizontal adaptation, when an article uses another as a methodological template to apply computational frameworks to independent datasets for validation across different research contexts, and iv) cross-pollination, when articles aggregate and synthesize data or methods from multiple sources to generate novel insights that transcend individual publications.
The reuse patterns transform fragmented scientific publishing into what Knuth envisioned as true composability, where small, well-understood computational and narrative components combine to form increasingly sophisticated knowledge structures.28 Such interlacing enables researchers working transparently and methodically to construct robust foundations for discovery, with each publication serving as both standalone contribution and building block for future work.
In conclusion, woven literature converts the culmination of scientific record from a static collection of isolated PDFs into a dynamic, interconnected (i.e., federated) infrastructure where cumulative knowledge emerges through systematic reuse and extension of validated components. The following subsection introduces open-source solutions and community efforts making interconnected, executable scholarship a practical reality.
Open-source pathways to realizing woven literature
NeuroLibre29 represents one of the earliest fully open-source implementations aligned with the principles of woven literature: https://neurolibre.org. Developed under the Canadian Open Neuroscience Platform,30 it started as a full-fledged reproducible publishing platform for neuroscience by integrating the open-source applications by JOSS31 with the documentation engines of the Jupyter Project.32 It currently hosts 19 next-gen preprints mostly in the fields of neuroimaging and MRI engineering, with additional submissions actively coming in as the platform moves toward its second iteration: https://evidencepub.io.
Each NeuroLibre publication is deeply interwoven with its reproducibility artifacts, which are automatically archived on Zenodo (https://zenodo.org) and served on demand through interactive computational environments. Nearly 60GB of input data is made readily accessible within these containerized reproducible runtimes, enabling real-time exploration and interactivity.
Other notable efforts toward modernizing scientific publishing include eLife’s Executable Research Articles33 and Distill (https://distill.pub),34 both of which are no longer actively maintained. Wholetale (https://wholetale.org)35 offered another model for stitching together reproducibility assets into executable articles, providing a limited set of example publications. Quarto (https://quarto.org) is an open-source scientific and technical publishing system that supports R, Python, Julia, and JavaScript, allowing users to produce documents, websites, and presentations that combine narrative with executable code. Observable (https://observablehq.com) provides a reactive notebook environment based in JavaScript, well-suited for building rich, interactive data visualizations and exploratory narratives, though its primary use has been outside formal scientific publishing. In parallel, Curvenote (https://curvenote.com)36 has made substantial contributions to next-gen publishing workflows, particularly through its development of Markedly Structured Text (MyST) markdown language,37 a foundational technology for bringing about the woven literature. Another open-source demonstration of MyST’s potential came with you-only-write-thrice, a proof-of-concept workflow that enabled publishing the same content across multiple formats.38
While these initiatives differ in openness, scope, and long-term sustainability, NeuroLibre stands out as an actively growing, fully open, and community-driven infrastructure grounded in the principles of open science and reproducibility. Its publication workflow has been made possible by a thriving ecosystem of open-source projects, whose ongoing innovations have not only enabled its core functionality but also steadily expanded its capabilities.
The sections below provide a brief review of existing tools that support each thread of a next-gen article, including reproducible runtimes, code and data integration, and editorial management systems that orchestrate the publication process.
Reproducible runtime
One of the main building blocks of NeuroLibre is BinderHub, a cloud-native platform that enables users to share reproducible, interactive computing environments directly from code repositories.39,40 Binder streamlines the process of creating containerized environments by reducing complex Dockerfile specifications to a small set of configuration files interpreted by language-agnostic buildpacks. This simplicity, combined with its seamless integration with Jupyter Notebooks, has made Binder one of the most widely adopted tools for executable research. As of 2025, the public Binder infrastructure (https://mybinder.org) serves up to 12,000 daily sessions across thousands of repositories (https://archive.analytics.mybinder.org), demonstrating its pivotal role in lowering the barrier to reproducible, in-browser scientific computing.
Google Colab is another widely used platform for interactive computing, particularly popular in data science and machine learning education.41 It allows users to write and execute Python code in a browser-based Jupyter Notebook interface backed by Google’s cloud infrastructure. Colab’s ease of use, integration with Google Drive, and free access to GPU and TPU resources have made it especially attractive for rapid prototyping, tutorials, and sharing notebooks across diverse user groups.
However, while both BinderHub and Colab offer interactive computational environments, they differ significantly in design philosophy and sustainability model. BinderHub is grounded in open science principles and supports community-led infrastructure. A notable example is 2i2c (https://2i2c.org), a nonprofit organization that operates scalable BinderHub services for research and education, exemplifying how interactive computing can be delivered sustainably through a community-driven approach. In contrast, Colab is a freemium product provided by a commercial entity, with priorities shaped by business models rather than academic interests.
BinderHub is fully open source and purpose-built to support reproducible science, emphasizing long-term preservation and transparency. It leverages the Reproducible Execution Environment Specification (REES),42 which uses configuration files like environment.yml, requirements.txt, or install.R to explicitly declare and version-control runtime dependencies. These specifications are used to build Docker images that can be hosted in private registries and archived, providing a robust foundation for sustained reproducibility. This model aligns strongly with the goals of scientific publishing, where preserving the integrity and reusability of computational environments is essential.
In contrast, Colab’s environments are transient and subject to changes in the underlying base images, which may lead to inconsistencies in results over time. While Colab offers generous access to GPU hardware out of the box (at least as of 2025), BinderHub deployments can be configured to integrate with GPU-enabled Kubernetes clusters or JupyterHub spawners, offering comparable computational capabilities within an infrastructure designed for reproducibility and archival.
An emerging alternative leverages WebAssembly,43 allowing code execution directly in the browser without relying on a remote server. Pyodide, a Python distribution compiled to WebAssembly, enables this shift by running Python code client-side in a manner similar to JavaScript.44 JupyterLite builds on Pyodide to offer a lightweight, serverless Jupyter environment that launches instantly in the browser.45 Here, the user interacts with notebooks whose code is executed locally, eliminating the need for backend infrastructure.
This approach is particularly exciting for the future of interactive scientific publishing. It offers a scalable solution for delivering interactive papers without being constrained by centralized compute resources, making it ideal for publications with modest computational demands and minimal data dependencies. It opens the door to creating rich, Python-based interactive articles akin to those pioneered by https://distill.pub, without requiring expertise in web-native programming. However, ensuring long-term preservation of these browser-executed environments remains an open challenge that warrants further exploration in the context of scholarly publishing.
Code and prose
A wide range of open-source tools now support the creation of executable scientific documents, enabling researchers to combine code and scientific prose in a single environment for reproducible and interactive outputs. R Markdown, introduced in 2012, laid the groundwork for integrating analysis and narrative, allowing for dynamic document generation across various formats.46 Two years later, GitBook (https://www.gitbook.com) emerged, popularizing structured, book-like web documentation, albeit not specifically tailored for scientific communication and currently evolved into a hosted commercial platform. In 2016, Bookdown extended R Markdown’s capabilities, facilitating the creation of books and long-form reports with features like cross-referencing and multi-format output.47
The landscape of structured, git-backed web content was bridged with the Jupyter ecosystem by JupyterBook,37 introduced in 2019 as part of the Executable Books (EB) (https://executablebooks.org). Leveraging Sphinx (https://www.sphinx-doc.org), a Python-based documentation engine, and MyST markdown language, it renders Jupyter Notebooks and Markdown files into interactive, publication-grade documents as static HTML files. Features such as collapsible code cells, interactive plotting, and live code execution via Thebe48 enhance its suitability for reproducible and reader-friendly content.
The MyST markdown language has been a major driver of the EB’s evolution, enabling its documentation engine to expand beyond a Sphinx-based static site generator toward a framework capable of serving documents as dynamic web pages. This transition (see more here) was supported through collaboration with Curvenote, which contributed a TypeScript library that leverages the mdast package from the UnifiedJS project (https://unifiedjs.com) to represent the entire content of Jupyter Notebooks and Markdown as abstract syntax trees. Building on this development, MyST-MD (https://mystmd.org) enables the full content of an article to be expressed as structured data (Figure 1c), supporting programmatic access and seamless transformation into a wide range of themes (e.g., single-page or book-like web layouts) and formats (e.g., PDF, XML, JATS, MECA, DOCX, or LaTeX) for publishing.
This structured data representation not only eliminates the need for complex format conversions in the publishing pipeline but also unlocks richer online functionality. It enables actionable content reuse, transclusion, and semantic understanding of elements such as figures, narrative text, and code cells. As such, MyST-MD represents a significant milestone in the realization of woven literature, and is currently supported by NeuroLibre for dynamically publishing living preprints.
Data
One of the most commonly missing pieces in building a next-gen article is the input data required for executable content to generate expected outputs. During its early development, the Binder project recognized this challenge and initially recommended storing datasets directly within GitHub repositories.39 However, this was acknowledged as a stopgap measure rather than a viable long-term strategy, given GitHub’s limitations as a platform designed primarily for version-controlling source code. Instead, the Binder team advocated for more robust approaches like Dat,49 a distributed protocol for data synchronization and versioning.
To understand the importance of thoughtful data distribution in scientific publishing, it is useful to reflect on how digital media became widely shareable. Technologies like BitTorrent50 revolutionized peer-to-peer file sharing by allowing users to download large files in small pieces from multiple sources, making the process more efficient and resilient. However, while BitTorrent excels at distributing static content, it offers limited support for collaboration or tracking how files evolve, capabilities that are essential for reproducible science.
Version control systems such as Git introduced structured mechanisms for tracking changes in source code across distributed teams. These same ideas have inspired tools for managing datasets, especially in contexts where traceability and reproducibility are essential. For example, git-annex extends Git to support versioning of large files without storing them directly in the repository. DataLad (https://www.datalad.org),51 built on top of git and git-annex, is widely used in neuroscience for jointly managing code, data, and the links between them. It enables reproducible research workflows that track both inputs and outputs across time and collaborators.
Complementing these solutions, NeuroBagel (https://neurobagel.org) tackles a different but equally critical problem: metadata standardization and dataset interoperability. By harmonizing dataset descriptors, NeuroBagel enables federated queries across distributed datasets—making it easier for researchers to discover and access compatible data resources.52
A more general-purpose approach to distributed, versioned data is the InterPlanetary File System (IPFS).53 Like Dat,49 IPFS identifies files using content hashes, ensuring that a given address always resolves to the same version of a file. Based on peer-to-peer protocols similar to BitTorrent, IPFS can serve as a decentralized backbone for hosting static datasets and artifacts. This content-addressable architecture is particularly valuable in reproducible science, where exact snapshots of datasets must be preserved and referenced across time.
Another approach, particularly relevant to scientific computing infrastructures, is the CERN Virtual Machine File System (CVMFS).54 Originally developed to distribute software in high-energy physics, CVMFS is well-suited to delivering large-scale datasets efficiently across distributed systems, with support for secure repositories. Its caching mechanisms and content-addressable design complement reproducibility goals by ensuring users access consistent versions of data, even at scale.
For platforms like NeuroLibre, which deploys executable scientific articles on JupyterHub instances running atop Kubernetes, mounting datasets to individual user sessions introduces unique challenges. Each reader session requires read-only access to the same inputs to ensure consistent reproducibility, while also being isolated and ephemeral.
To address this, NeuroLibre recognizes and supports data hosted on repositories like Zenodo, OSF, and DataLad, using a tool called repo2data to automatically fetch and cache these inputs in a reproducible manner. This makes it possible to reproduce results from linked datasets without requiring manual setup. Looking ahead, the next phase of NeuroLibre’s infrastructure will adopt more scalable, efficient solutions such as CVMFS, paving the way for a more robust and larger ecosystem of executable research objects.
Editorial management and peer review
While advances in tools for code, data, and runtime integration have made it more technically feasible than ever to move beyond the BOOMER paradigm, infrastructure alone does not ensure reproducibility. Without careful curation and modern editorial oversight, executable content written in notebooks may still fall short of yielding truly reproducible outputs.55
To address this gap, NeuroLibre pioneered the integration of a technical screening process into the living preprint publication workflow. This process supports curation by establishing an iterative interaction between the submitting author, a technical screener, and an editorial bot (RoboNeuro) on a GitHub issue. Within this workflow, the runtime environment is verified, reproducibility checks are logged, and any necessary changes are communicated and resolved until the submission passes all technical criteria (see examples here).
The platform and tooling that support this next-gen technical screening workflow were built from a fork of the Open Journals’ applications, Buffy and JOSS,7 and were extended with additional components designed to interface with NeuroLibre APIs. Unlike the JOSS workflow, which involves peer review of both the scientific content and the functionality of the associated research software, NeuroLibre’s technical screening focuses solely on ensuring that computational outputs are generated as expected. It does not involve scientific evaluation, similar to commonly used preprint services such as arXiv.org that has posted more than 1 million preprints over the last three decades.56
Interestingly, preprints have not only made publicly funded research more accessible, but have also spearheaded the development of new approaches to peer-review, such as eLife’s “publish, then review” model.57 This shift reflects a broader recognition that review should not be a one-time gatekeeping event as in the BOOMER paradigm, but an ongoing, distributed process that involves diverse perspectives and evolves alongside the work itself. It is on this ground that new models and infrastructures are beginning to take shape, aiming to support more transparent, inclusive, and sustained forms of scholarly evaluation.
A key contributor to advancing this procedural transformation has been the Confederation of Open Access Repositories (COAR), which supports the development of aligned open access infrastructure through a global network of universities, funders, and research institutions.58,59 One of its notable initiatives, COAR Notify, proposes a decentralized mechanism to connect research outputs hosted in distributed repositories with external services (such as overlay journals and open peer review platforms) using linked-data notifications.60 This effort stands at the intersection of numerous preprint servers (e.g., arXiv, bioRxiv, medRxiv, Zenodo, Research Square, SciFLO, and Center of Open Science, to name a few), and more importantly, preprint review initiatives such as PreReview (https://prereview.org), Peer Community in (PCI, https://peercommunityin.org), eLife (https://elifesciences.org), Plaudit (https://plaudit.pub), SciPost (https://scipost.org), and Peeref (https://www.peeref.com). Additionally, platforms like ResearchHub (https://researchhub.com) are pioneering novel approaches to post-publication peer review through decentralized evaluation mechanisms and introducing ResearchCoin as an innovative initiative to shift incentive structures in favor of researchers. Collectively, these initiatives represent a growing movement to build a more open, interoperable, and participatory scholarly communication ecosystem where peer review and evaluation are no longer tied to the traditional boundaries of legacy publishers.61
Results
Figure 3 highlights the structural shift from conventional publishing to a reproducible research article. On the left, the static PDF presents frozen text, figures, and code snippets, which offer limited transparency or reusability.62 On the right, the same study is rendered as a next-gen article on NeuroLibre, where narrative prose is stitched together with executable code, openly accessible data, and an embedded runtime.62 This format allows readers to interact with the actual software package and learn about its applications in real-time with zero installation. The NiMARE paper is one of 19 living preprints hosted on NeuroLibre that illustrate this paradigm in practice: https://neurolibre.org.
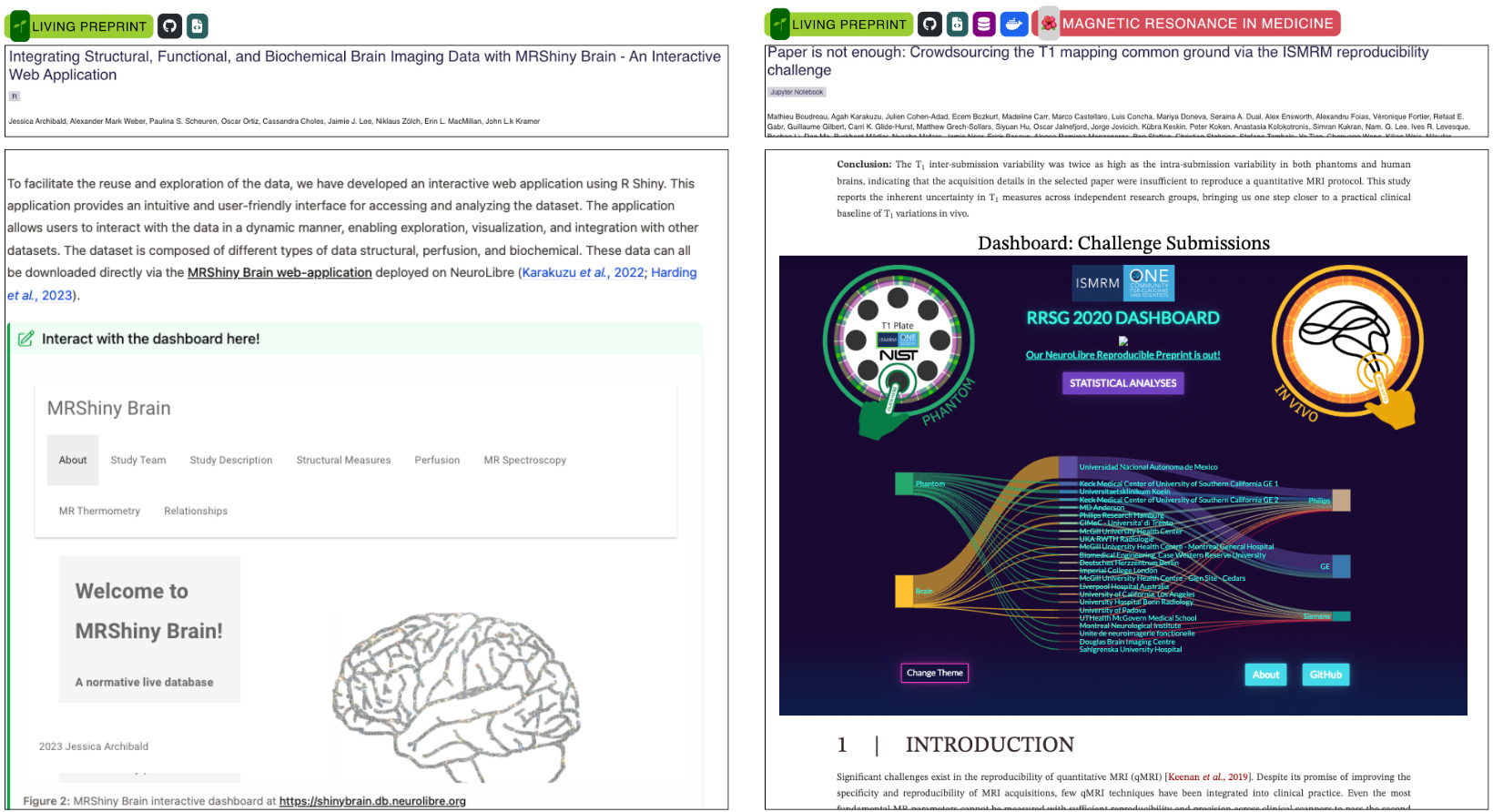
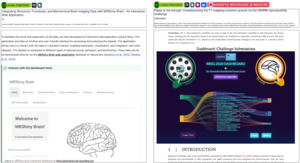
Figure 4 showcases how next-gen publishing on NeuroLibre enables the seamless integration of interactive dashboards as first-class figures. The MRShiny Brain dashboard (https://shinybrain.db.neurolibre.org) offers an interface for exploring structural, perfusion, and metabolic brain imaging data from a normative cohort.63 Built using R Shiny, it empowers readers to navigate multidimensional outcome measures, such as metabolite concentrations and MR thermometry, without being constrained by static layouts or figure limits.
On the right, a Plotly Dash dashboard supports the ISMRM T1 Mapping Reproducibility Challenge, visualizing inter- and intra-site variability across phantom and human scans (https://rrsg2020.db.neurolibre.org). This web-native figure offers toggles, filters, and drill-down visualizations that would require dozens or even hundreds of static panels in a conventional PDF. These dashboards are deployed on NeuroLibre’s dedicated Dokku (https://dokku.com) deployment for serving interactive data applications, underscoring the platform’s commitment to supporting researchers motivated to communicate complex results through novel, reader-driven interfaces.
Reuse patterns enabled by federated publishing networks demonstrate their transformative potential for scientific validation and knowledge synthesis. Longitudinal extension is exemplified by,64 where the publication evolved continuously through data submissions from multiple sites participating in a reproducibility challenge. The meta-analysis template provided in65 illustrates the horizontal adaptation pattern, as it was subsequently reused by66 with an entirely different dataset, enabling systematic validation of methodological robustness across research contexts. Cross-pollination, as defined earlier, can be seen in a proof of concept implementation at https://agahkarakuzu.github.io/paperception, yet no real-world examples currently exist.
Discussion
Adoption of next-gen publishing falters from incentives, not interfaces
Despite the maturity and accessibility of technologies enabling next-gen articles, their adoption in mainstream scientific publishing remains limited. The culprit is not an insurmountable learning curve that prevents the broader adoption of such tools. If anything, for the next generation researchers, using tools like Binder and Jupyter Notebooks is as natural as word processors were for those trained in the BOOMER ecosystem. It is worth bearing in mind that this new cohort of scientists has grown up with smartphones and now navigates information fluently using LLMs. For them, the tools of woven literature feel intuitive in a landscape where coding for end-user programming is becoming less a specialized skill and more a facet of AI literacy. In this context, a gen-alpha researcher would aptly describe their fluency as “vibe coding straight through those interactive plots into something reproducible, no sweat”.
The real obstacles lie elsewhere. For publishers, legacy business models tied to static formats continue to dominate, offering little incentive to adopt infrastructures that would undercut their profit margins. On the researchers’ side, entrenched incentive structures and network effects still reward publication in “high-impact” journals, regardless of the reproducibility or interactivity of the work. Addressing these issues requires not just advocacy, but the development and visibility of alternative venues that demonstrate what modern scholarly communication can look like. Founded in 2025, the Continuous Science Foundation (https://continuousfoundation.org) exemplifies this shift through a community-driven approach that places sustainability at its core.
An additional and often overlooked barrier is the difficulty of sustaining platforms like NeuroLibre through traditional scientific funding mechanisms. Despite their potential to transform research communication and reproducibility at scale, infrastructure projects of this nature often fall outside the scope of conventional grant criteria. While programs such as the Chan Zuckerberg Initiative’s open science funding (https://chanzuckerberg.com) provide crucial support, there remains a need for broader awareness and recognition within public and national funding systems.
Investments in open, reproducible publishing infrastructure may appear modest in size, but their long-term return in terms of saved time, avoided duplication, and enhanced scientific rigor is substantial. Building sustainable pathways for funding these efforts is essential to realizing the full promise of networked science publishing.67
Defining the edge where a scientific workflow transitions into a next-gen publication
The computational and data demands of next-gen articles vary significantly across research domains. For instance, neuroimaging studies often involve processing large volumes of high-resolution MRI data combined with advanced statistical and machine learning methods. Meanwhile, theoretical papers typically have lower data needs but may require intensive symbolic computation. In contrast, fields such as qualitative social sciences often involve relatively small datasets and minimal computational processing, focusing instead on interpretive analysis.
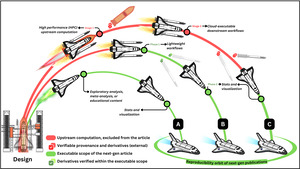
Figure 5 illustrates how scientific workflows enter the reproducibility orbit of next-gen publications by transitioning from external computation stages to internal executable phases. The degree to which a workflow enters this orbit depends on its complexity and structure. Some workflows are fully self-contained, with all operations reproducible in a single session, while others involve modular segments that depend on externally computed stages or pre-generated artifacts that are generated internally.
-
Trajectory A represents lightweight use cases that do not require batch processing or multi-phase workflow execution logic. All computations, from data ingestion to analysis and visualization, are included within the article and can be executed independently, without strict ordering or dependencies between notebooks. Readers can interact with any part of the content asynchronously, reproducing results in one go without needing to trace interdependent phases.
-
Trajectory B represents multiple interdependent phases that are all technically reproducible within the available computational limits, but differ in practicality for reader interaction. The earlier phases, such as data cleaning, simple model training, or transformation etc., may discourage readers from re-running them interactively. However, these phases are still included in the publication and can be executed if desired. To streamline the reading experience, pre-executed outputs from these initial phases are verified (green checkmarks) and provided as derivatives. Readers are encouraged to engage with the later, more interactive phases, such as analysis, visualization, or interpretation, without being required to reproduce the full pipeline from scratch. This model maintains full transparency and reproducibility, while accommodating asynchronous entry points aligned with reader expectations.
-
Trajectory C reflects computationally intensive workflows whose upstream stages exceed the runtime or resource constraints of next-gen publishing platforms. These workflows often involve multi-step pipelines, such as large-scale HPC preprocessing, simulation, or training procedures, that must be executed externally before publication. Only the final phase, typically involving analysis, visualization, or interpretive synthesis, is included within the executable scope of the next-gen article.
For example, NeuroLibre currently allocates up to 3GB of RAM and 1 CPU per living print user session, with support for storing up to 4GB of input data. These limits are often sufficient to meet the computational requirements of Trajectory A (Figure 5). Representative examples include: i) data-driven exploratory analyses, such as a living preprint by Bellec et al.68 showcasing a word feature analysis using scikit-learn,69 ii) a science communication analysis for interactive exploration of patent history,70 iii) interactive tutorials covering neuroimaging meta-analyses62 and quantitative MRI,71,72 and iv) interactive meta-analyses performed focused on myelin imaging65 and brain imaging demographics in Quebec.66
An example representative of Trajectory B is the living preprint resulting from a multi-center reproducibility challenge organized by the International Society for Magnetic Resonance in Medicine (ISMRM), which evaluated the inter-site reliability of T1 mapping.64,73 In this case, the initial computational phase, such as aligning region-of-interest (ROI) masks and fitting T1 maps, was decoupled from the subsequent statistical analyses, which could be executed within a few minutes.
An example of Trajectory C can be found in a living preprint by Wang et al.,74 which benchmarks the reproducibility of resting-state fMRI denoising strategies across varying preprocessing pipelines. The upstream processing required to generate these results involved running multiple fMRI pipelines under different parameter configurations, an operation that demanded high-performance computing resources beyond the scope of a typical interactive article. As such, only the downstream derivatives, including statistical analysis and visualization of the precomputed outputs, were integrated into the living preprint. This allowed readers to interact with and explore the results reproducibly, even though the computationally intensive preprocessing steps had to be completed externally.
Seamless continuation from upstream workflows to reproducible publishing
Integrating outputs from upstream stages into next-gen articles in a reliable and reproducible way remains an open challenge. Fortunately, platforms like NeuroDesk have begun to address this gap by providing portable, containerized environments capable of running complex neuroimaging workflows.75 NeuroDesk enables reproducible execution and captures the provenance between inputs and outputs, ensuring traceability across computational stages. Similarly, CBRAIN provides a web-based interface to high-performance computing resources, enabling researchers to run large-scale data analyses while preserving metadata and provenance.76 Another example is BrainLife, focusing on reproducible neuroscience pipelines, offering cloud-based, interactive execution of modular workflows.77 More recently, O2S2PARC was introduced as a cloud-based platform with a partial pay-per-use model to support the execution and sharing of multiscale computational models in life sciences.78
An integration between NeuroLibre and such platforms could offer a seamless continuation from heavy upstream processing to interactive, downstream communication, bridging the full research workflow that can extend from the study inception to publication.79,80 Emerging infrastructures, such as Orvium (https://orvium.io), and DeSci Nodes (https://nodes.desci.com), further open possibilities for verifiable, tamper-proof records of these computational steps on blockchain, encouraging new models of trust and attribution in scholarly publishing.81,82
Conclusion
The ultimate convergence of technical and procedural publishing innovations reviewed in this article will mark a critical turning point in scholarly communication. By integrating reproducibility-aware open infrastructure with distributed models of peer review, scholarly publishing is approaching the replacement of BOOMER’s static, one-shot logic with the continuous, collaborative, and verifiable attributes of the woven literature.
A Turkish expression, “ilmek ilmek dokumak” literally means “to weave stitch by stitch” but it is used metaphorically to describe doing something with great care, patience, and attention to detail. This captures the spirit of woven literature: a vision of scholarly publishing built meticulously by interlacing code, data, prose, and peer review into a coherent whole, one next-gen article at a time.
As we move forward, this stitch-by-stitch approach offers not just a more reproducible and transparent future, but also one that values the craft of knowledge creation itself, a quality more vital than ever in an era shaped by the rapid rise of generative AI and automated content.
The threads are already in motion; what lies ahead is ours to weave. We can choose to remain entangled in the static logic of the BOOMER paradigm, or co-create a knowledge network that evolves through novel reuse patterns, modularity, and reproducibility.
Acknowledgements
I thank Nikola Stikov for insightful discussions, for reading the initial draft of this manuscript, and for providing thoughtful feedback. I am also grateful to the members of the Canadian Open Neuroscience Platform (CONP) Publishing Committee and to the researchers who contributed to the funding and development of NeuroLibre, including Elizabeth DuPre, Lune Bellec, Jean-Baptiste Poline, Mathieu Boudreau, Patrick Bermudez, Samir Das, and Alan C. Evans. Special thanks to Rowan Cockett and Chris Holdgraf, as well as all members of the Executable Books and Project Jupyter communities for their unwavering commitment to improving the tools that unlocked the networked science publishing.
Funding was received from the CONP (https://conp.ca), Brain Canada (https://braincanada.ca), Courtois Neuromod (https://www.cneuromod.ca), Quebec Bio-imaging Network (QBIN), and the Canada First Research Excellence Fund (TransMedTech).
Conflicts of Interest
The author declares no conflicts of interest.


_and_a_next-gen_reproducible_art.png)


_and_a_next-gen_reproducible_art.png)