Recent research has shown that continuous and naturalistic visual stimuli, such as narrative movies, can evoke brain responses in high-level visual regions that are stable for timescales on the order of seconds to tens of seconds.1 What drives these temporal dynamics in the brain? One possible explanation is that high-level sensory regions have inherently slower dynamics, which are present even in the absence of a stimulus2 and in infants.3 Having slow-changing representations allows for the accumulation of information over time, providing context for interpreting the current stimulus based on information from the recent past.4,5
An alternative explanation for the long-timescale stability of high-level sensory regions could be that these temporal dynamics are, at least in part, due to dynamics inherent to the stimulus itself. Recent work by Heusser et al.6 sought to quantify the dynamics of content in a narrative movie by using topic modeling and hidden Markov models (HMMs) to discretize continuous stimulus into events characterized by their trajectories through semantic space. They found that the movie itself exhibited stable semantic events, suggesting that the event structure in brain responses could be “inherited” from the temporal structure of the stimulus, rather than arising from slow cortical dynamics. However, this approach relies on human annotations of stimulus content, rather than being directly derived from the stimulus alone.
Our approach to better understand this long-timescale stability in high-level sensory regions is to characterize the dynamics in brain responses driven purely by stimulus, using a frame-by-frame prediction model for visual cortex. Here we introduce a new python package, img2fmri, to predict group-level fMRI responses to individual images. This prediction model uses an artificial deep neural network (DNN), as DNNs have been successful at predicting cortical responses in the human visual cortex when trained on real world visual categorization tasks.7 These neural networks learn to extract features (e.g. shapes, textures, eyes) from naturalistic visual data that allow them to accurately classify objects, animals, and scenes in the images they process,8 and can also be used to extract those predominant features from input to subsequently be used in predicting cortical responses.9 Research has also shown that the hierarchy of layers in a trained DNN can predict along a hierarchy of processing in the brain, where deeper, or higher, layers in a DNN best predict higher levels of cortical processing.10,11 Our model is built by combining a pretrained ResNet-18 DNN with a linear regression model to predict fMRI responses to individual images. The mapping from DNN to the brain is fit using data from the open source BOLD5000 project,12 which includes fMRI responses for three subjects viewing 4916 unique images drawn from ImageNet,13 COCO,14 and SUN.15 For each image, we predict activity patterns for five visual regions of interest (ROIs) for each subject’s brain (defined by the BOLD5000 project): an early visual region of voxels near the calcarine sulcus sensitive to visual stimuli (EarlyVis), the lateral occipital complex (LOC), an object-selective region,16 as well as the occipital place area (OPA), retrosplenial cortex (RSC), and parahippocampal place area (PPA), three scene-selective regions.17
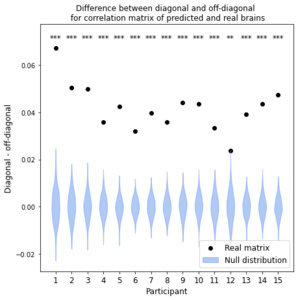
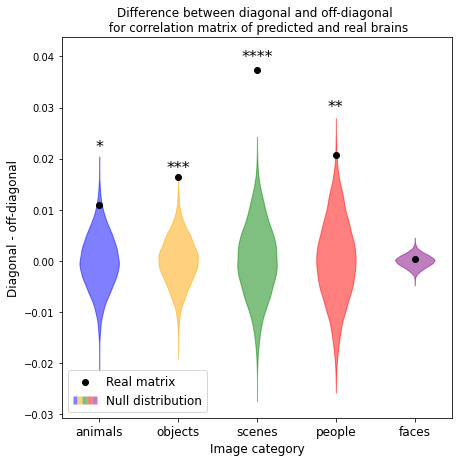
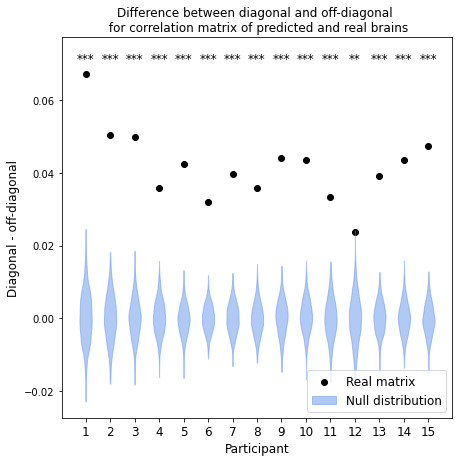
To validate our model, we predict fMRI responses to images our model has not previously seen from the Twinset dataset.18 Our model can significantly predict fMRI responses at the group level for most categories in this dataset (Fig 1), and can significantly predict responses in every individual subject (Fig 2) († p<0.10, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001).
We then show how our frame-by-frame prediction model can be extended to a continuous visual stimulus by predicting an fMRI response to Pixar Animation Studio’s short film Partly Cloudy. Here we compare the timepoint-timepoint similarity from our predicted frame-by-frame response to the timepoint-timepoint similarity in the actual group-averaged fMRI response, using data from the 33 adults in Richardson et al.19 For comparison, we also attempted to predict brain dynamics using a baseline model based only on the low-level luminance of the visual stimulus. In analyzing the timepoint-timepoint similarity of our predicted fMRI response around human-annotated event boundaries in the movie (Fig 3), we find that our model outperforms the luminance model in describing the dynamics of the real fMRI response around these event boundaries, particularly in the timepoints just before an event, and at an event itself (* p<0.05, ** p<0.01). These analyses suggest that in visual areas of the brain, at least some of the temporal dynamics we see in the brain’s processing of continuous, naturalistic stimuli can be explained by dynamics in the stimulus itself, since they can be predicted from our frame-by-frame model.
All analyses, notebooks, and code can be found at https://github.com/dpmlab/img2fmri. The README in this repository outlines installation steps of background software, with the primary requirements being Python 3.9 or higher, PyTorch, and neuroimaging softwares AFNI and FSL. We also include a Dockerfile and Docker image (via Docker Hub) for a pre-installed container. Our package is being released under the MIT license, and is also released as a pip/PyPI package, with API documentation available on ReadTheDocs.
To use the model and view these analyses, we encourage readers to explore our overview.ipynb in the previously linked GitHub repository. For more information on the training of our model using the open source BOLD5000 dataset and pretrained ResNet-18 DNN, we have included a notebook model_training.ipynb within our model_training folder that outlines the model training process and offers suggestions for extending the model to predict fMRI responses from other feature-detecting models, and to other brain ROIs. Users can report any issues via GitHub issues, as outlined in CONTRIBUTING.rst, or by emailing the authors at mbb2176@columbia.edu.